Abstract
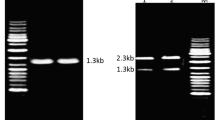
A gene encoding an extracellular protease from Dichelobacter nodosus was characterized and expressed in E. coli rosetta-gami (DE3). The nucleotide sequence analysis revealed an ORF of 1427 bp ecoding 475 amino acids long protein of calculated molecular weight 50.6 kDa and pI value 6.09. The phylogenetic analysis showed relatedness to subtilisin-like serine proteases of peptidase S8 family. The amino acid sequence analysis showed presence of N-terminal pre-peptide (1–23 aa), pro-peptide (24–160 aa), peptidase S8 domain (161–457 aa), and a C-terminal extension (458–475 aa). The gene harboring native signal peptide was expressed in pET-22b(+) for production of AprV2 recombinant protein. SDS-PAGE revealed the highest production of IPTG induced recombinant protein ∼37 kDa at 16 °C after 16 h. The purified protein after Ni-NTA affinity chromatography showed single protein band of ∼37 kDa which was also confirmed by the detection of blue coloured band of same size in Western blotting. The recombinant protein showed activity over broad temperature and pH range with optimum at 35 °C and pH 7.0. Similarly, the enzyme was stable over broad range 15–65 °C and 4–10 pH with maximum stability at 25 °C and pH 6. The activity of purified enzyme was also stimulated in the presence of Ca2+. The purified enzyme showed highest activity towards casein as compared to gelatin and BSA. These findings suggest AprV2 as an important candidate for industrial applications such as pharmaceuticals.





Similar content being viewed by others
References
Bennett, G. N., & Hickford, J. G. H. (2011). Ovine footrot: New approaches to an old disease. Veterinary Microbiology, 148, 1–7.
Kennan, R. M., Gilhuus, M., Frosth, S., Seemann, T., Dhungyel, O. P., Whittington, R. J., Boyce, J. D., Powell, D. R., Aspan, A., Jorgensen, H. J., Bulach, D. M., & Rooda, J. I. (2014). Genomic evidence for a globally distributed. Bimodal Population in the Ovine Footrot Pathogen Dichelobacter nodosus. mBio, 5(5), e01821–14.
Egerton, J. R., Roberts, D. S., & Parsonson, I. M. (1969). The aetiology and pathogenesis of ovine footrotA histological study of the bacterial invasion. Journal of Comparative Pathology, 79, 209–216.
Stewart, D. J., Clark, B. L., & Jarret, R. G. (1984). Difference between strains of Bacteroides nodosus in their effects on the severity of footrot, bodyweight and wool growth on Merino sheep. Australian Veterinary Journal, 61, 349–352.
Ghosh, A., Chakrabarti, K., & Chattopadhyay, D. (2009). Cloning of feather-degrading minor extracellular protease from Bacillus cereus DCUW: dissection of the structural domains. Microbiology, 155, 2049–57.
Arnorsdottir, J., Smaradottir, R. B., Magnusson, O. T., Thorbjarnardottir, S. H., Eggertsson, G., & Kristjansson, M. (2002). Characterization of a cloned subtilisin-like serine proteinase from a psychrotrophic Vibrio species. European Journal of Biochemistry, 269, 5536–5546.
Kwon, K., Hasseman, J., Latham, S., Grose, C., Do, Y., Fleischmann, R. D., Pieper, R., & Petersen, S. N. (2011). Recombinant expression and functional analysis of proteases from Streptococcus pneumoniae, Bacillus anthracis and Yersinia pestis. BMC Biochemistry, 12, 17.
Larsen, A. N., Moe, E., Helland, R., Gjellesvik, D. R., & Willassen, N. P. (2006). Characterization of a recombinantly expressed proteinase K-like enzyme from a psychrotrophic Serratia sp. FEBS Journal, 273, 47–60.
Maurer, K. H. (2004). Detergent proteases. Current Opinion in Biotechnology, 15, 330–334.
Craik, C. S., Page, M. J., & Madison, E. L. (2011). Proteases as therapeutics. Biochemical Journal, 435, 1–16.
Chanalia, P., Gandhi, D., Jodha, D., & Singh, J. (2011). Applications of microbial proteases in pharmaceutical industry: an overview. Reviews in Medical Microbiology, 22, 96–101.
Shen, H. B., & Chou, K. C. (2009). Identification of proteases and their types. Analytical Biochemistry, 385, 153–160.
Skerman, T. M. (1975). Determination of some in vitro growth requirements of Bacteroides nodosus. Journal of General Microbiology, 87, 107–119.
La Fontaine, S., Egerton, J. R., & Rood, J. I. (1993). Detection of Dichelobacter nodosus using species specific oligonucleotides as PCR primers. Veterinary Microbiology, 35, 101–117.
Dhungyel, O. P., Whittington, R. J., & Egerton, J. R. (2002). Serogroup specific single and multiplex PCR with pre-enrichment culture and immuno-magnetic bead capture for identifying strains of D. nodosus in sheep with footrot prior to vaccination. Molecular and Cellular Probes, 16, 285–296.
Cheetham, B. F., Tanjung, L. R., Sutherland, M., Druitt, J., Green, G., McFarlane, J., Bailey, G. D., Seaman, J. T., & Katz, M. E. (2006). Improved diagnosis of virulent footrot using intA gene. Veterinary Microbiology, 116, 166–174.
Palmer, M. A. (1993). A gelatin test to detect activity and stability of proteases produced by Dichelobacter (Bacteroides) nodosus. Veterinary Microbiology, 36, 113–122.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410.
Letunic, C. R. R., Pils, B., Pinkert, S., Schultz, J., & Bork, P. (2006). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Research, 34, D257–260.
Marchler-Bauer, A., Anderson, J. B., Cherukuri, P. F., DeWeese-Scott, C., Geer, L. Y., He, M. G. S., Hurwitz, D. I., Jackson, J. D., Ke, Z., Lanczycki, C. J., Liebert, C. A., Liu, C., Lu, F., Marchler, G. H., Mullokandov, M., Shoemaker, B. A., Simonyan, V., Song, J. S., Thiessen, P. A., Yamashita, R. A., Yin, J. J., Zhang, D., & Bryant, S. H. (2005). CDD: a Conserved Domain Database for protein classification. Nucleic Acids Research, 33, 192–196.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739.
Schwede, T., Kopp, J., Guex, N., & Peitsch, M. C. (2003). SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Research, 31, 3381–3385.
Benkert, P., Tosatto, S. C. E., & Schomburg, D. (2008). QMEAN: A comprehensive scoring function for model quality assessment. Proteins: Structure, Function, and Bioinformatics, 71(1), 261–277.
Folin, O., & Ciocalteu, V. (1927). On tyrosine and tryptophane determinations in proteins. Journal of Biological Chemistry, 73, 627–650.
Wang, Q., Miao, J. L., Hou, Y. H., Ding, Y., Wang, G. D., & Li, G. Y. (2005). Purification and characterization of an extracellular cold–active serine protease from the psychrophilic bacterium Colwellia sp. NJ341. Biotechnology Letters, 27, 1195–1198.
Rao, C. S., Sathish, T., Brahamaiah, P., Kumar, T. P., & Prakasham, R. S. (2009). Development of a mathematical model for Bacillus circulans growth and alkaline protease production kinetics. Journal of Chemical Technology and Biotechnology, 84, 302–307.
Kennan, R. M., Wong, W., Dhungyel, O. P., Han, X., Wong, D., Parker, D., Rosado, C. J., Law, R. H., McGowan, S., Reeve, S. B., Levina, V., Powers, G. A., Pike, R. N., Bottomley, S. P., Smith, A. I., Marsh, I., Whittington, R. J., Whisstock, J. C., Porter, C. J., & Rood, J. I. (2010). The Subtilisin-like protease AprV2 is required for virulence and uses a novel disulphide-tethered exosite to bind substrates. PLoS Pathogens, 6(11), E1001210.
Myers, G. S., Parker, D., Al-Hasani, K., Kennan, R. M., Seemann, T., Ren, Q., Badger, J. H., Selengut, J. D., Deboy, R. T., Tettelin, H., Boyce, J. D., McCarl, V. P., Han, X., Nelson, W. C., Madupu, R., Mohamoud, Y., Holley, T., Fedorova, N., Khouri, H., Bottomley, S. P., Whittington, R. J., Adler, B., Songer, J. G., Rood, J. I., & Paulsen, I. T. (2007). Genome sequence and identification of candidate vaccine antigens from the animal pathogen Dichelobacter nodosus. Nature Biotechnology, 25(5), 569–575.
Riffkin, M. C., Wang, L. F., Kortt, A. A., & Stewart, D. J. (1995). A single amino-acid change between the antigenically different extracellular serine proteases V2 and B2 from Dichelobacter. Gene, 167(1–2), 279–283.
Deng, A., Wu, J., Zhang, G., & Wen, T. (2011). Molecular and structural characterization of a surfactant-stable high-alkaline protease AprB with a novel structural feature unique to subtilisin family. Biochimie, 93, 783–791.
VanderLaan, J. M., Teplyakov, A. V., Kelders, H., Kalk, K. H., Misset, O., Mulleners, L. J., & Dijkstra, B. W. (1992). Crystal structure of the high-alkaline serine protease PB92 from Bacillus alcalophilus. Protein Engineering, 5(5), 405–411.
Wandersman, C. (1989). Secretion, processing and activation of bacterial extracellular proteases. Molecular Microbiology, 3(12), 1825–1831.
Wong, W., Kennan, R. M., Rosado, C. J., Rood, J. I., Whisstocka, J. C., & Portera, C. J. (2010). Crystallization of the virulent and benign subtilisin-like proteases from the ovine footrot pathogen Dichelobacter nodosus. Acta Crystallographica Section, F66, 289–293.
Rawlings, N. D., & Barett, A. J. (1994). Families of serine peptidases. Methods in Enzymology, 224, 19–61.
Ather, A. (2009). Identification, cloning and expressions of proteases from a cold adapted organism Aliivibrio salmonicida KJE-3900. University of Tromso.
Vaughan, P. R., Wang, L. F., Stewart, D. J., Lilley, G. G., & Kortt, A. A. (1994). Expression in Escherichia coli of the extracellular basic protease frorn Dichelobacter nodosus. Microbiology, 140, 2093–2100.
Sarvas, M., Harwood, C. R., Bron, S., & Dijl, J. M. (2004). Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochimica et Biophysica Acta, 1694, 311–327.
Godde, C., Sahm, K., Brouns, S. J. J., Kluskens, L. D., Oost, J. V., deVos, W. M., & Antranikian, G. (2005). Cloning and expression of islandisin, a new thermostable subtilisin from Fervidobacterium islandicum, in Escherichia coli. Applied and Environmental Microbiology, 71(7), 3951–3958.
Chen, X. L., Xie, B. B., Lu, J. T., He, H. L., & Zhang, Y. Z. (2007). A novel type of subtilase from the psychrotolerant bacterium Pseudoalteromonas sp. SM9913: catalytic and structural properties of deseasin MCP-01. Microbiology, 153, 2116–2125.
Huston, A. L., Methe, B., & Deming, J. W. (2004). Purification, characterization, sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Applied and Environmental Microbiology, 70, 2321–2328.
Kortt, A. A., Caldwell, J. B., Lilley, G. G., Edwards, R., Vaughan, J., & Stewart, D. J. (1994). Characterization of a basic serine protease (pI ∼ 9.5) secreted by virulent strains of Dichelobacter nodosus and identification of a distinct, but closely related, proteinase secreted by benign strains. Journal of Biochemistry, 299, 521–525.
Setyorini, E., Kim, Y. J., Takenaka, S., Murakami, S., & Aoki, K. (2006). Purification and characterization of a halotolerant intracellular protease from Bacillus subtilis strain FP-133. Journal of Basic Microbiology, 46(4), 284–304.
Yan, B. Q., Chen, X. L., Hou, X. Y., He, H., Zhou, B. C., & Zhang, Y. Z. (2009). Molecular analysis of the gene encoding a cold-adapted halophilic subtilase from deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913: cloning, expression, characterization and function analysis of the C-terminal PPC domains. Extremophiles, 13, 725–733.
Pushpam, P. L., Rajesh, T., & Gunasekaran, P. (2011). Identification and characterization of alkaline serine protease from goat skin surface metagenome. AMB Express, 1, 3.
Zhang, Y., Zhao, J., & Zeng, R. (2011). Expression and characterization of a novel mesophilic protease from metagenomic library derived from Antarctic coastal sediment. Extremophiles, 15, 23–29.
Doddapaneni, K. K., Tatineni, R., Potumarthi, R., & Mangamoori, L. N. (2007). Optimization of media constituents through response surface methodology for improved production of alkaline proteases by Serratia rubidaea. Journal of Chemical Technology and Biotechnology, 82, 721–729.
Morita, Y., Hasan, Q., Sakaguchi, T., Murakami, Y., Yokoyama, K., & Tamiya, E. (1998). Properties of a cold-active protease from psychrotrophic Flavobacterium balustinum P104. Applied Microbiology and Biotechnology, 50(6), 669–675.
Secades, P., Alvarez, B., & Guijarro, J. A. (2001). Purification and characterization of a psychrophilic calcium induced, growth-phase dependent metalloprotease from the fish pathogen Flavobacterium psychrophilum. Applied and Environmental Microbiology, 67(6), 2436–2444.
Tariq, A. L., Reyaz, A. L., & Prabakaran, J. J. (2011). Purification and Characterization of 56 KDa cold active protease from Serratia marcescens. African Journal of Microbiology Research, 5(32), 5841–5847.
Shankar, S., Rao, M., & Laxman, R. S. (2011). Purification and characterization of an alkaline protease by a new strain of Beauveria sp. Process Biochemistry, 46, 579–585.
Srilakshmi, J., Madhavi, J., Lavanya, S., & Ammani, K. (2015). Commercial potential of fungal protease: past, present and future prospects. Journal of Pharmaceutical, Chemical and Biological Sciences, 2(4), 218–234.
Acknowledgments
The authors acknowledge the financial support received from the ICAR under NAIP project on footrot. The authors also acknowledge CSK-Himachal Pradesh Agricultural University, Palampur, for logistic support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Wani, A.H., Sharma, M., Salwan, R. et al. Cloning, Expression, and Functional Characterization of Serine Protease Aprv2 from Virulent Isolate Dichelobacter nodosus of Indian Origin. Appl Biochem Biotechnol 180, 576–587 (2016). https://doi.org/10.1007/s12010-016-2117-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2117-5