Abstract
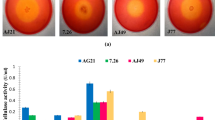
High costs of natural cellulose utilization and cellulase production are an industrial challenge. In view of this, an isolated soil actinobacterium identified as Promicromonospora sp. VP111 showed potential for production of major cellulases (CMCase, FPase, and β-glucosidase) utilizing untreated agricultural lignocellulosic wastes. Extensive disintegration of microcrystalline cellulose and adherence on it during fermentation divulged true cellulolytic efficiency of the strain. Conventional optimization resulted in increased cellulase yield in a cost-effective medium, and the central composite design (CCD) analysis revealed cellulase production to be limited by cellulose and ammonium sulfate. Cellulase activities were enhanced by Co+2 (1 mM) and retained up to 60 °C and pH 9.0, indicating thermo-alkaline tolerance. Cellulases showed stability in organic solvents (25 % v/v) with log P ow ≥ 1.24. Untreated wheat straw during submerged fermentation was particularly degraded and yielded about twofold higher levels of cellulases than with commercial cellulose (Na-CMC and avicel) which is especially economical. Thus, this is the first detailed report on cellulases from an efficient strain of Promicromonospora that was non-hemolytic, alkali-halotolerant, antibiotic (erythromycin, kanamycin, rifampicin, cefaclor, ceftazidime) resistant, multiple heavy metal (Mo+6 = W+6 > Pb+2 > Mn+2 > Cr+3 > Sn+2), and organic solvent (n-hexane, isooctane) tolerant, which is industrially and environmentally valuable.

Similar content being viewed by others
References
Tanimura, A., Liu, W., Yamada, K., Kishida, T., & Toyohara, H. (2013). Animal cellulases with a focus on aquatic invertebrates. Fisheries Science, 79, 1–13.
Béguin, P., & Aubert, J. P. (1994). The biological degradation of cellulose. FEMS Microbiology Reviews, 13, 25–58.
Bhat, M. K. (2000). Cellulases and related enzymes in biotechnology. Biotechnology Advances, 18, 355–383.
Wilson, D. B. (2009). Cellulases and biofuels. Current Opinion in Biotechnology, 20, 295–299.
Phitsuwan, P., Laohakunjit, N., Kerdchoechuen, O., Kyu, K. L., & Ratanakhanokchai, K. (2013). Present and potential applications of cellulases in agriculture, biotechnology, and bioenergy. Folia Microbiologica, 58, 163–176.
Acebal, C., Castillon, P., Estrada, P., Mata, I., Costa, E., Aguado, J., Romero, D., & Jimenez, F. (1986). Enhanced cellulase production from Trichoderma reesei QM 9414 on physically treated wheat straw. Applied Microbiology and Biotechnology, 24, 218–223.
Matkar, K., Chapla, D., Divecha, J., Nighojkar, A., & Madamwar, D. (2013). Production of cellulase by a newly isolated strain of Aspergillus sydowii and its optimization under submerged fermentation. International Biodeterioration and Biodegradation, 78, 24–33.
Trivedi, N., Gupta, V., Kumar, M., Kumari, P., Reddy, C. R. K., & Jha, B. (2011). Solvent tolerant marine bacterium Bacillus aquimaris secreting organic solvent stable alkaline cellulase. Chemosphere, 83, 706–712.
Asha, B. M., & Sakthivel, N. (2014). Production, purification and characterization of a new cellulase from Bacillus subtilis that exhibit halophilic, alkaliphilic and solvent-tolerant properties. Annals of Microbiology, 64, 1839–1848.
Doukyu, N., & Ogino, H. (2010). Organic solvent tolerant enzymes. Biochemical Engineering Journal, 48, 270–282.
Bisht, D., Yadav, S. K., Gautam, P., & Darmwal, N. S. (2013). Simultaneous production of alkaline lipase and protease by antibiotic and heavy metal tolerant Pseudomonas aeruginosa. Journal of Basic Microbiology, 53, 715–722.
Rahman, Z., & Singh, V. P. (2014). CrVI reduction by Enterobacter sp. DU17 isolated from the tannery waste dump site and characterization of the bacterium and the CrVI reductase. International Biodeterioration and Biodegradation, 91, 97–103.
Sadhu, S., Ghosh, P. K., De, T. K., & Maiti, T. K. (2013). Optimization of cultural condition and synergistic effect of lactose with carboxymethyl cellulose on cellulase production by Bacillus sp. isolated from fecal matter of elephant Elephas maximus. Advances in Microbiology, 3, 280–288.
Saha, S., Roy, R. J., Sen, S. K., & Ray, A. K. (2006). Characterization of cellulase-producing bacteria from the digestive tract of tilapia, Oreochromis mossambica Peters and grass carp, Ctenopharyngodon idella Valenciennes. Aquaculture Research, 37, 380–388.
Gao, W., Lee, E. J., Lee, S. U., Li, J., Chung, C. H., & Lee, J. W. (2012). Enhanced carboxymethylcellulase production by a newly isolated marine bacterium Cellulophaga lytica LBH-14, using wheat bran. Journal of Microbiology and Biotechnology, 22, 1412–1422.
Lo, Y. C., Saratale, G. D., Chen, W. M., Bai, M. D., & Chang, J. S. (2009). Isolation of cellulose-hydrolytic bacteria and applications of the cellulolytic enzymes for biohydrogen production. Enzyme and Microbial Technology, 44, 417–425.
Balasubramanian, N., Toubarro, D., Teixeira, M., & Simõs, N. (2012). Purification and biochemical characterization of a novel thermo-stable carboxymethyl cellulase from Azorean isolate Bacillus mycoides S122C. Applied Biochemistry and Biotechnology, 168, 2191–2204.
Amore, A., Pepe, O., Ventorino, V., Birolo, L., Giangrande, C., & Faraco, V. (2013). Industrial waste based compost as a source of novel cellulolytic strains and enzymes. FEMS Microbiology Letters, 339, 93–101.
Teather, R. M., & Wood, P. J. (1982). Use of congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Applied and Environmental Microbiology, 43, 777–780.
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., & Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proceedings of the National Academy of Sciences of the United States of America, 82, 6955–6959.
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., Park, S. C., Jeon, Y. S., Lee, J. H., Yi, H., Won, S., & Chun, J. (2012). Introducing EzTaxon-e a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. International Journal of Systematic and Evolutionary Microbiology, 62, 716–721.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6 molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Plackett, R. L., & Burman, J. P. (1946). The design of optimum multifactorial experiments. Biometrika, 33, 305–325.
Weuster-Botz, D. (2000). Experimental design for fermentation media development statistical design or global random search? Journal of Bioscience and Bioengineering, 90, 473–483.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428.
Wood, T. M., & Bhat, K. M. (1998). Method for measuring cellulase activities. In W. A. Wood & J. A. Kellogg (Eds.), Methods in enzymology (pp. 87–112). New York: Academic.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Chevallet, M., Luche, S., & Rabilloud, T. (2006). Silver staining of proteins in polyacrylamide gels. Nature Protocols, 1, 1852–1858.
McCarthy, A. J. (1987). Lignocellulose-degrading actinomycetes. FEMS Microbiology Reviews, 46, 145–163.
Lynd, L. R., Weimer, P. J., van Zyl, W. H., & Pretorius, I. S. (2002). Microbial cellulose utilization fundamentals and biotechnology. Microbiology and Molecular Biology Reviews, 66, 506–577.
Anderson, I., Abt, B., Lykidis, A., Klenk, H. P., Kyrpides, N., & Ivanova, N. (2012). Genomics of aerobic cellulose utilization systems in actinobacteria. PloS One, 7, e39331.
Kang, S. M., Khan, A. L., Hamayun, M., Hussain, J., Joo, G. J., You, Y. H., Kim, J. G., & Lee, I. J. (2012). Gibberellin-producing Promicromonospora sp. SE188 improves Solanum lycopersicum plant growth and influences endogenous plant hormones. The Journal of Microbiology, 50, 902–909.
Kumar, M., Joshi, A., Kashyapa, R., & Khannab, S. (2011). Production of xylanase by Promicromonospora sp. MARS with rice straw under non sterile conditions. Process Biochemistry, 46, 1614–1618.
Qin, S., Jiang, J. H., Klenk, H. P., Zhu, W. Y., Zhao, G. Z., Zhao, L. X., Tang, S. K., Xu, L. H., & Li, W. J. (2012). Promicromonospora xylanilytica sp. nov., an endophytic actinomycete isolated from surface-sterilized leaves of the medicinal plant Maytenus austroyunnanensis. International Journal of Systematic and Evolutionary Microbiology, 62, 84–89.
Ventorino, V., Aliberti, A., Faraco, V., Robertiello, A., Giacobbe, S., Ercolini, D., Amore, A., Fagnano, M., & Pepe, O. (2015). Exploring the microbiota dynamics related to vegetable biomasses degradation and study of lignocellulose-degrading bacteria for industrial biotechnological application. Scientific Reports, 5, 8161.
Deswal, D., Khasa, Y. P., & Kuhad, R. C. (2011). Optimization of cellulase production by a brown rot fungus Fomitopsis sp. RCK2010 under solid state fermentation. Bioresource Technology, 102, 6065–6072.
Yoko, I. (1986). Effect of glycine and its derivatives on production and release of β-galactosidase by Escherichia coli. Agricultural and Biological Chemistry, 50, 2747–2753.
Dhawan, S., & Kuhad, R. C. (2002). Effect of amino acids and vitamins on laccase production from bird’s nest fungus Cyathus bulleri. Bioresource Technology, 84, 35–38.
Spiridonov, N. A., & Wilson, D. B. (1998). Regulation of biosynthesis of individual cellulases in Thermomonospora fusca. Journal of Bacteriology, 180, 3529–3532.
Rajoka, M. I. (2004). Influence of various fermentation variables on exoglucanase production in Cellulomonas flavigena. Electronic Journal of Biotechnology, 7, 259–266.
Asha, B. M., Revathi, M., Yadav, A., & Sakthivel, N. (2012). Purification and characterization of a thermophilic cellulase from a novel cellulolytic strain, Paenibacillus barcinonensis. Journal of Microbiology and Biotechnology, 22, 1501–1509.
Saxena, S., Bahadur, J., & Varma, A. (1992). Effect of cobalt and nickel on growth and carboxymethyl cellulase activity of Cellulomonas spp. BioMetals, 5, 209–212.
Ferchak, J. D., & Pye, E. K. (1983). Effect of cellobiose, glucose, ethanol, and metal ions on the cellulase enzyme complex of Thermomonospora fusca. Biotechnology and Bioengineering, 25, 2865–2872.
Zaks, A., & Klibanov, A. M. (1988). Enzymatic catalysis in nonaqueous solvents. The Journal of Biological Chemistry, 263, 3194–3201.
Li, X., Wang, H. L., Li, T., & Yu, H. Y. (2012). Purification and characterization of an organic solvent-tolerant alkaline cellulase from a halophilic isolate of Thalassobacillus. Biotechnology Letters, 34, 1531–1536.
Ogino, H., & Ishikawa, H. (2001). Enzymes which are stable in the presence of organic solvents. Journal of Bioscience and Bioengineering, 91, 109–116.
Li, X., & Yu, H. Y. (2012). Purification and characterization of an organic solvent-tolerant cellulase from a halotolerant isolate, Bacillus sp. L1. The Journal of Industrial Microbiology and Biotechnology, 39, 1117–1124.
Li, X., & Yu, H. Y. (2013). Halostable cellulase with organic solvent tolerance from Haloarcula sp. LLSG7 and its application in bioethanol fermentation using agricultural wastes. The Journal of Industrial Microbiology and Biotechnology, 40, 1357–1365.
Saratale, G. D., Kshirsagar, S. D., Sampange, V. T., Saratale, R. G., Oh, S. E., Govindwar, S. P., & Oh, M. K. (2014). Cellulolytic enzymes production by utilizing agricultural wastes under solid state fermentation and its application for biohydrogen production. Applied Biochemistry and Biotechnology, 174, 2801–2817.
Chellapandi, P., & Jani, H. M. (2008). Production of endoglucanase by the native strains of Streptomyces isolates in submerged fermentation. The Brazilian Journal of Microbiology, 39, 122–127.
Saratale, G. D., Saratale, R. G., & Oh, S. E. (2012). Production and characterization of multiple cellulolytic enzymes by isolated Streptomyces sp. MDS. Biomass and Bioenergy, 47, 302–315.
de Menezes, A. B., Lockhart, R. J., Cox, M. J., Allison, H. E., & McCarthy, A. J. (2008). Cellulose degradation by micromonosporas recovered from freshwater lakes and classification of these actinomycetes by DNA Gyrase B gene sequencing. Applied and Environmental Microbiology, 74, 7080–7084.
Waldron, C. R., Jr., Becker-Vallone, C. A., & Eveleigh, D. E. (1986). Isolation and characterization of a cellulolytic actinomycete Microbispora bispora. Applied Microbiology and Biotechnology, 24, 477–486.
Saratale, G. D., & Oh, S. E. (2011). Production of thermotolerant and alkalotolerant cellulolytic enzymes by isolated Nocardiopsis sp. KNU. Biodegradation, 22, 905–919.
Acknowledgments
Fellowships from C.S.I.R. (New Delhi) to Lebin Thomas, from U.G.C. (New Delhi) to Hari Ram and Alok Kumar, and R&D grant from the University of Delhi are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
(DOC 488 kb)
Supplementary Fig. 2
(DOC 197 kb)
Supplementary Fig. 3
(DOC 251 kb)
Supplementary Fig. 4
(DOC 1858 kb)
Supplementary Fig. 5
(DOC 232 kb)
Supplementary Fig. 6
(DOC 539 kb)
Supplementary Table 1
(DOC 35 kb)
Supplementary Table 2
(DOC 34 kb)
Supplementary Table 3
(DOC 35 kb)
Supplementary Table 4
(DOC 36 kb)
Supplementary Table 5
(DOC 36 kb)
Supplementary Table 6
(DOC 37 kb)
Supplementary Table 7
(DOC 38 kb)
Supplementary Table 8
(DOC 35 kb)
Rights and permissions
About this article
Cite this article
Thomas, L., Ram, H., Kumar, A. et al. Production, Optimization, and Characterization of Organic Solvent Tolerant Cellulases from a Lignocellulosic Waste-Degrading Actinobacterium, Promicromonospora sp. VP111. Appl Biochem Biotechnol 179, 863–879 (2016). https://doi.org/10.1007/s12010-016-2036-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2036-5