Abstract
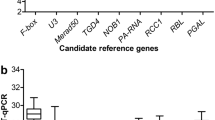
Brinjal/eggplant/aubergine is one of the major solanaceous vegetable crops. Recent availability of genome information greatly facilitates the fundamental research on brinjal. Gene expression patterns during different stages of fruit development can provide clues towards the understanding of its biological functions. Quantitative real-time PCR (qPCR) has become one of the most widely used methods for rapid and accurate quantification of gene expression. However, its success depends on the use of a suitable reference gene for data normalization. For qPCR analysis, a single reference gene is not universally suitable for all experiments. Therefore, reference gene validation is a crucial step. Suitable reference genes for qPCR analysis of brinjal fruit development have not been investigated so far. In this study, we have selected 21 candidate reference genes from the Brinjal (Solanum melongena) Plant Gene Indices database (compbio.dfci.harvard.edu/tgi/plant.html) and studied their expression profiles by qPCR during six different fruit developmental stages (0, 5, 10, 20, 30, and 50 days post anthesis) along with leaf samples of the Pusa Purple Long (PPL) variety. To evaluate the stability of gene expression, geNorm and NormFinder analytical softwares were used. geNorm identified SAND (SAND family protein) and TBP (TATA binding protein) as the best pairs of reference genes in brinjal fruit development. The results showed that for brinjal fruit development, individual or a combination of reference genes should be selected for data normalization. NormFinder identified Expressed gene (expressed sequence) as the best single reference gene in brinjal fruit development. In this study, we have identified and validated for the first time reference genes to provide accurate transcript normalization and quantification at various fruit developmental stages of brinjal which can also be useful for gene expression studies in other Solanaceae plant species.





Similar content being viewed by others
References
Collonnier, C., Fock, I., Kashyap, V., Rotino, G. L., Daunay, M. C., Lian, Y., et al. (2001). Applications of biotechnology in eggplant. Plant Cell Tissue and Organ Culture, 65, 91–107. doi:10.1023/A: 1010674425536.
Doganlar, S., Frary, A., Daunay, M. C., Lester, R. N., & Tanksley, S. D. (2002). A comparative genetic linkage map of eggplant (Solanum melongena L.) and its implications for genome evolution in the Solanaceae. Genetics, 161, 1697–1711.
Kumar, G., Meena, B. L., Kar, R., Tiwari, S. K., Gangopadhyay, K. K., Bisht, I. S., et al. (2008). Morphological diversity in brinjal (Solanum melongena L.) germplasm accessions. Characterization and utilization. Plant Genetic Resources: Characterization and Utilization, 6(3), 232–236. doi:10.1017/S1479262108994211.
Nunome, T., Negoro, S., Kono, I., Kanamori, H., Miyatake, K., Yamaguchi, H., et al. (2009). Development of SSR markers derived from SSR-enriched genomic library of eggplant (Solanum melongena L.). Theoretical and Applied Genetics, 119, 1143–1153. doi:10.1007/s00122-009-1116-0.
Fukuoka, H., Yamaguchi, H., Nunome, T., Negoro, S., Miyatake, K., & Ohyama, A. (2010). Accumulation, functional annotation, and comparative analysis of expressed sequence tags in eggplant (Solanum melongena L.), the third pole of the genus Solanum species after tomato and potato. Gene, 450, 76–84. doi:10.1016/j.gene.2009.10.006.
Polignano, G., Uggenti, P., Bisignano, V., & Gatta, C. D. (2010). Genetic divergence analysis in eggplant (Solanum melongena L.) and allied species. Genetic Resources and Crop Evolution, 57, 171–181. doi:10.1007/s10722-009-9459-6.
Barchi, L., Lanteri, S., Portis, E., Acquadro, A., Vale, G., Toppino, L., et al. (2011). Identification of SNP and SSR markers in eggplant using RAD tag sequencing. BMC Genomics, 12, 304. doi:10.1186/1471-2164-12-304.
Rieu, I., & Powers, S. J. (2009). Real-time quantitative RT-PCR: design, calculations and statistics. The Plant Cell, 21, 1031–1033. doi:10.1105/tpc.109.066001.
Guenin, S., Mauriat, M., Pelloux, J., Wuytswinkel, O. V., Bellini, C., & Gutierrez, L. (2009). Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. Journal of Experimental Botany, 60(2), 487–493. doi:10.1093/jxb/ern305.
Le, D. T., Aldrich, D. L., Valliyodan, B., Watanabe, Y., Ha, C. V., Nishiyama, R., et al. (2012). Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS ONE, 7(9), e46487. doi:10.1371/journal.pone.0046487.
Imai, T., Ubi, B. E., Saito, T., & Moriguchi, T. (2014). Evaluation of reference genes for accurate normalization of gene expression for real-time quantitative PCR in Pyrus pyrifolia using different tissue samples and seasonal conditions. PLoS ONE, 9(1), e86492. doi:10.1371/journal.pone.0086492.
Bustin, S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. Journal of Molecular Endocrinology, 29, 23–39. doi:10.1677/jme.0.0290023.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Roy, N. V., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3(7), 0034.1–0034.11. doi:10.1186/gb-2002-3-7-research0034.
De Almeida, M. R., Ruedell, C. M., Ricachenevsky, F. K., Sperotto, R. A., Pasquali, G., & Fett-Neto, A. G. (2010). Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill. BMC Molecular Biology, 11, 73. doi:10.1186/1471-2199-11-73.
Gachon, C., Mingam, A., & Charrier, B. (2004). Real-time PCR: what relevance to plant studies? Journal of Experimental Botany, 55(402), 1445–1454. doi:10.1093/jxb/erh181.
Nolan, T., Hands, R. E., & Bustin, S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nature Protocols, 1(3), 1559–1582. doi:10.1038/nprot.2006.236.
Kubista, M., Andrade, J. M., Bengtsson, M., Forootan, A., Jonak, E. J., Lind, K., et al. (2006). The real-time polymerase chain reaction. Molecular Aspects of Medicine, 27, 95–125. doi:10.1016/j.mam.2005.12.007.
Cassan-Wang, H., Soler, M., Yu, H., Camargo, E. L. O., Carocha, V., Ladouce, N., et al. (2012). Reference genes for high-throughput quantitative reverse transcription–PCR analysis of gene expression in organs and tissues of Eucalyptus grown in various environmental conditions. Plant and Cell Physiology, 53(12), 2101–2116. doi:10.1093/pcp/pcs152.
Yeap, W.-C., Loo, J. M., Wong, Y. C., & Kulaveerasingam, H. (2014). Evaluation of suitable reference genes for qRT-PCR gene expression normalization in reproductive, vegetative tissues and during fruit development in oil palm. Plant Cell Tissue Organ Culture, 116, 55–66. doi:10.1007/s11240-013-0382-3.
Dheda, K., Huggett, J. F., Chang, J. S., Kima, L. U., Bustinc, S. A., Johnson, M. A., et al. (2005). The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Analytical Biochemistry, 344, 141–143. doi:10.1016/j.ab.2005.05.022.
Gutierrez, L., Mauriat, M., Guenin, S., Pelloux, J., Lefebvre, J. F., Louvet, R., et al. (2008). The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnology Journal, 6, 609–618. doi:10.1111/j.1467-7652.2008.00346.x. Epub 2008 Apr 22.
Ferguson, B. S., Nam, H., Hopkins, R. G., & Morrison, R. F. (2010). Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in adipocytes. PLoS ONE, 5(12), e15208. doi:10.1371/journal.pone.0015208.
Kong, Q., Yuan, J., Gao, L., Zhao, S., Jiang, W., Huang, Y., et al. (2014). Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS ONE, 9(2), e90612. doi:10.1371/journal.pone.0090612.
Watson, J. D., Hopkins, N. H., Roberts, J. W., Steitz, J. A., & Weiner, A. M. (1965). The functioning of higher eukaryotic genes. Molecular Biology of the Gene, 1(21), 704.
Warrington, J. A., Nair, A., Mahadevappa, M., & Tsyganskaya, M. (2000). Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiological Genomics, 2, 143–147.
Silver, N., Best, S., Jiang, J., & Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology, 7, 33. doi:10.1186/1471-2199-7-33.
Kouadjo, K. E., Nishida, Y., Cadrin-Girard, J. F., Yoshioka, M., & St-Amand, J. (2007). Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics, 8, 127. doi:10.1186/1471-2164-8-127.
Andersen, C. L., Jensen, J. L., & Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250. doi:10.1158/0008-5472.
Exposito-Rodriguez, M., Borges, A. A., Borges-Perez, A., & Perez, J. A. (2008). Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biology, 8, 131. doi:10.1186/1471-2229-8-131.
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., & Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Molecular Biology, 10, 11. doi:10.1186/1471-2199-10-11.
Hellemans, J., Mortier, G., Paepe, A. D., Speleman, F., & Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology, 8(2), R19. doi:10.1186/gb-2007-8-2-r19.
Gantasala, N. P., Papolu, P. K., Thakur, P. K., Kamaraju, D., Sreevathsa, R., & Rao, U. (2013). Selection and validation of reference genes for quantitative gene expression studies by real-time PCR in eggplant (Solanum melongena L). BMC Research Notes, 6, 312. doi:10.1186/1756-0500-6-312.
Carles, C. C., & Fletcher, J. C. (2009). The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes and Development, 23, 2723–2728. doi:10.1101/gad.1812609.
Mafra, V., Kubo, K. S., Alves-Ferreira, M., Ribeiro-Alves, M., Stuart, R. M., Boava, L. P., et al. (2012). Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS ONE, 7(2), e31263. doi:10.1371/journal.pone.0031263.
Nakano, T., Fujisawa, M., Shima, Y., & Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. Journal of Experimental Botany, 65(12), 3111–9. doi:10.1093/jxb/eru154.
Demidenko, N. V., Logacheva, M. D., & Penin, A. A. (2011). Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS ONE, 6(5), e19434. doi:10.1371/journal.pone.0019434.
Zhu, X., Li, X., Chen, W., Chen, J., Lu, W., Chen, L., et al. (2012). Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS ONE, 7(8), e44405. doi:10.1371/journal.pone.0044405.
Padmalatha, K. V., Dhandapani, G., Kanakachari, M., Kumar, S., Dass, A., Patil, D. P., et al. (2012). Genome-wide transcriptomic analysis of cotton under drought stress reveals significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Molecular Biology, 78, 223–246. doi:10.1007/s11103-011-9857-y.
Artico, S., Nardeli, S. M., Brilhante, O., Grossi-de-Sa, M. F., & Alves-Ferreira, M. (2010). Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biology, 10, 49. doi:10.1186/1471-2229-10-49.
LiLi, T., XianLong, Z., DiQiu, L., ShuangXia, J., JingLin, C., LongFu, Z., et al. (2007). Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chinese Science Bulletin, 52(22), 3110–3117. doi:10.1007/s11434-007-0461-0.
Wan, H., Yuan, W., Ruan, M., Ye, Q., Wang, R., Li, Z., et al. (2011). Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochemical and Biophysical Research Communications, 416, 24–30. doi:10.1016/j.bbrc.2011.10.105.
Jian, B., Liu, B., Bi, Y., Hou, W., Wu, C., & Han, T. (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Molecular Biology, 9, 59. doi:10.1186/1471-2199-9-59.
Schmidt, G. W., & Delaney, S. K. (2010). Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Molecular Genetics and Genomics, 283, 233–241. doi:10.1007/s00438-010-0511-1.
Reddy, D. S., Bhatnagar-Mathur, P., Cindhuri, K. S., & Sharma, K. K. (2013). Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut. PLoS ONE, 8(10), e78555. doi:10.1371/journal.pone.0078555.
Barsalobres-Cavallari, C., Severino, F. E., Maluf, M. P., & Maia, I. G. (2009). Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Molecular Biology, 10, 1. doi:10.1186/1471-2199-10-1.
Chandna, R., Augustine, R., & Bisht, N. C. (2012). Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR. PLoS ONE, 7(5), e36918. doi:10.1371/journal.pone.0036918.
Amil-Ruiz, F., Garrido-Gala, J., Blanco-Portales, R., Folta, K. M., Munoz-Blanco, J., & Caballero, J. L. (2013). Identification and validation of reference genes for transcript normalization in strawberry (Fragaria ananassa) defense responses. PLoS ONE, 8(8), e70603. doi:10.1371/journal.pone.0070603.
Tong, Z., Gao, Z., Wang, F., Zhou, J., & Zhang, Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Molecular Biology, 10, 71. doi:10.1186/1471-2199-10-71.
Liavonchanka, A., & Feussner, I. (2006). Lipoxygenases: occurrence, functions and catalysis. Journal of Plant Physiology, 163, 348–357. doi:10.1016/j.jplph.2005.11.006.
Porta, H., & Rocha-Sosa, M. (2002). Plant lipoxygenases: physiological and molecular features. Journal of Plant Physiology, 130, 15–21. doi:10.1104/pp. 010787.
Feng, B., Dong, Z., Xu, Z., An, X., Qin, H., Wu, N., et al. (2010). Molecular analysis of lipoxygenase (LOX) genes in common wheat and phylogenetic investigation of LOX proteins from model and crop plants. Journal of Cereal Science, 52, 387–394. doi:10.1016/j.jcs.2010.06.019.
Umate, P. (2011). Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signaling & Behavior, 6(3), 335–338. doi:10.4161/psb.6.3.13546.
Wang, R., Shen, W., Liu, L., Jiang, L., Liu, Y., Su, N., et al. (2008). A novel lipoxygenase gene from developing rice seeds confers dual position specificity and responds to wounding and insect attack. Plant Molecular Biology, 66(4), 401–414. doi:10.1007/s11103-007-9278-0.
Vellosillo, T., Martinez, M., Lopez, M. A., Vicente, J., Cascon, T., Dolan, L., et al. (2007). Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell, 19, 831–846. doi:10.1105/tpc.106.046052.
Acknowledgments
This work was supported by funds from the Indian Council of Agricultural Research (ICAR), New Delhi. We thank Dr. M. L. V. Phanindra for reviewing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Mogilicherla Kanakachar and Amolkumar U. Solanke contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1
Strategy for the identification of reference genes for qPCR normalization in brinjal (Solanum melongena L.) fruit development stages. (PPTX 216 kb)
Supplementary material 2
Analysis for efficiency of qPCR primers. (PPTX 577 kb)
Supplementary material 3
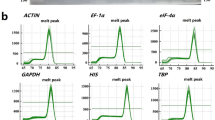
Real-time amplification specificity. Melt curves with a single peak generated for each of the 21 reference genes. (PPTX 1107 kb)
Supplementary material 4
Values of efficiency ± standard deviation (SD) of the primers for the housekeeping genes and average values of quantification cycle (Cq) ± standard deviation (SD) of biological replicates generated by the Miner to the reference genes of brinjal. (DOCX 16 kb)
Rights and permissions
About this article
Cite this article
Kanakachari, M., Solanke, A.U., Prabhakaran, N. et al. Evaluation of Suitable Reference Genes for Normalization of qPCR Gene Expression Studies in Brinjal (Solanum melongena L.) During Fruit Developmental Stages. Appl Biochem Biotechnol 178, 433–450 (2016). https://doi.org/10.1007/s12010-015-1884-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1884-8