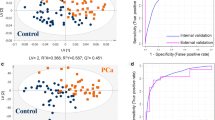
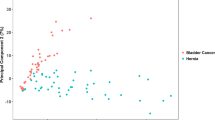
Abstract
Metabolomics is a post-genomics research field for analysis of low molecular weight compounds in biological samples and has shown great potentials for elucidating complex mechanisms associated with diseases. However, metabolomics studies on gastric cancer (GC), which is the second leading cause of cancer death worldwide, remain scarce, and the molecular mechanisms to metabolomics phenotypes are also still not fully understood. This study reports that the metabolic pathways can be exploited as biomarkers for diagnosis and treatment of GC progression as a case study. Importantly, the urinary metabolites and metabolic patterns were analyzed by high-throughput liquid chromatography mass spectrometry (LC-MS) metabolomics strategy coupled with chemometric evaluation. Sixteen metabolites (nine upregulated and seven downregulated) were differentially expressed and may thus serve as potential urinary biomarkers for human GC. These metabolites were mainly involved in multiple metabolic pathways, including citrate cycle (malic acid, succinic acid, 2-oxoglutarate, citric acid), cyanoamino acid metabolism (glycine, alanine), primary bile acid biosynthesis (glycine, taurine, glycocholic acid), arginine and proline metabolism (urea, l-proline), and fatty acid metabolism (hexadecanoic acid), among others. Network analysis validated close association between these identified metabolites and altered metabolic pathways in a variety of biological processes. These results suggest that urine metabolic profiles have great potential in detecting GC and may aid in understanding its underlying mechanisms. It provides insight into disease pathophysiology and can serve as the basis for developing disease biomarkers and therapeutic interventions for GC diseases.





Similar content being viewed by others
References
Hou, Q., Tan, H. T., Lim, K. H., Lim, T. K., Khoo, A., Tan, I. B., Yeoh, K. G., & Chung, M. C. (2013). Identification and functional validation of caldesmon as a potential gastric cancer metastasis-associated protein. Journal of Proteome Research, 12(2), 980–990.
Aquino, P. F., Fischer, J. S., Neves-Ferreira, A. G., Perales, J., Domont, G. B., Araujo, G. D., Barbosa, V. C., Viana, J., Chalub, S. R., Lima de Souza, A. Q., Carvalho, M. G., Leão de Souza, A. D., & Carvalho, P. C. (2012). Are gastric cancer resection margin proteomic profiles more similar to those from controls or tumors? Journal of Proteome Research, 11(12), 5836–5842.
Courant, F., Antignac, J. P., Monteau, F., & Le Bizec, B. (2013). Metabolomics as a potential new approach for investigating human reproductive disorders. Journal of Proteome Research, 12(6), 2914–2920.
Blazquez, M., Carretero, A., Ellis, J. K., Athersuch, T. J., Cavill, R., Ebbels, T. M., Keun, H. C., Castell, J. V., Lahoz, A., & Bort, R. (2013). A combination of transcriptomics and metabolomics uncovers enhanced bile acid biosynthesis in HepG2 cells expressing CCAAT/enhancer-binding protein β (C/EBPβ), hepatocyte nuclear factor 4α (HNF4α), and constitutive androstane receptor (CAR). Journal of Proteome Research, 12(6), 2732–2741.
Manna, S. K., Krausz, K. W., Bonzo, J. A., Idle, J. R., & Gonzalez, F. J. (2013). Metabolomics reveals aging-associated attenuation of noninvasive radiation biomarkers in mice: potential role of polyamine catabolism and incoherent DNA damage-repair. Journal of Proteome Research, 12(5), 2269–2281.
Hu, J. D., Tang, H. Q., Zhang, Q., Fan, J., Hong, J., Gu, J. Z., & Chen, J. L. (2011). Prediction of gastric cancer metastasis through urinary metabolomic investigation using GC/MS. World Journal of Gastroenterology, 17(6), 727–734.
Wang, X., Zhang, A., Han, Y., Wang, P., Sun, H., Song, G., Dong, T., Yuan, Y., Yuan, X., Zhang, M., Xie, N., Zhang, H., Dong, H., & Dong, W. (2012). Urine metabolomics analysis for biomarker discovery and detection of jaundice syndrome in patients with liver disease. Molecular & Cellular Proteomics, 11(8), 370–380.
Zhang, A., Sun, H., Han, Y., Yuan, Y., Wang, P., Song, G., Yuan, X., Zhang, M., Xie, N., & Wang, X. (2012). Exploratory urinary metabolic biomarkers and pathways using UPLC-Q-TOF-HDMS coupled with pattern recognition approach. Analyst, 137(18), 4200–4208.
Wang, X., Wang, H., Zhang, A., Lu, X., Sun, H., Dong, H., & Wang, P. (2012). Metabolomics study on the toxicity of aconite root and its processed products using ultraperformance liquid-chromatography/electrospray-ionization synapt high-definition mass spectrometry coupled with pattern recognition approach and ingenuity pathways analysis. Journal of Proteome Research, 11(2), 1284–1301.
Sun, H., Ni, B., Zhang, A., Wang, M., Dong, H., & Wang, X. (2012). Metabolomics study on fuzi and its processed products using ultra-performance liquid-chromatography/electrospray-ionization synapt high-definition mass spectrometry coupled with pattern recognition analysis. Analyst, 137(1), 170–185.
Wu, W.,, & Chung, M. C. (2013). The gastric fluid proteome as a potential source of gastric cancer biomarkers. Journal of Proteomics.
Wang, X., Yang, B., Sun, H., & Zhang, A. (2012). Pattern recognition approaches and computational systems tools for ultra performance liquid chromatography-mass spectrometry-based comprehensive metabolomic profiling and pathways analysis of biological data sets. Analytical Chemistry, 84(1), 428–439.
Yang, B., Zhang, A., Sun, H., Dong, W., Yan, G., Li, T., & Wang, X. (2012). Metabolomic study of insomnia and intervention effects of suanzaoren decoction using ultra-performance liquid-chromatography/electrospray-ionization synapt high-definition mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis, 58, 113–124.
Zhang, A., Sun, H., Wang, P., Han, Y., & Wang, X. (2012). Modern analytical techniques in metabolomics analysis. Analyst, 137(2), 293–300.
Dong, H., Zhang, A., Sun, H., Wang, H., Lu, X., Wang, M., Ni, B., & Wang, X. (2012). Ingenuity pathways analysis of urine metabolomics phenotypes toxicity of Chuanwu in Wistar rats by UPLC-Q-TOF-HDMS coupled with pattern recognition methods. Molecular BioSystems, 8(4), 1206–1221.
Zhang, A., Sun, H., Wang, P., Han, Y., & Wang, X. (2012). Recent and potential developments of biofluid analyses in metabolomics. Journal of Proteomics, 75(4), 1079–1088.
Wang, X., Sun, H., Zhang, A., Sun, W., Wang, P., & Wang, Z. (2011). Potential role of metabolomics approaches in the area of traditional Chinese medicine: as pillars of the bridge between Chinese and Western medicine. Journal of Pharmaceutical and Biomedical Analysis, 55(5), 859–868.
Wei, J., Xie, G., Ge, S., Qiu, Y., Liu, W., Lu, A., Chen, T., Li, H., Zhou, Z., & Jia, W. (2012). Metabolic transformation of DMBA-induced carcinogenesis and inhibitory effect of salvianolic acid b and breviscapine treatment. Journal of Proteome Research, 11(2), 1302–1316.
Zhang, A. H., Sun, H., Han, Y., Yan, G. L., Yuan, Y., Song, G. C., Yuan, X. X., Xie, N., & Wang, X. J. (2013). Ultraperformance liquid chromatography-mass spectrometry based comprehensive metabolomics combined with pattern recognition and network analysis methods for characterization of metabolites and metabolic pathways from biological data sets. Analytical Chemistry, 85, 7606–7612.
Zhang, A., Zhou, X., Zhao, H., Guan, Y., Zhou, S., Yan, G. L., Ma, Z., Liu, Q., & Wang, X. (2014). Rapidly improved determination of metabolites from biological data sets using the high-efficient TransOmics tool. Molecular BioSystems, 10, 160–165.
Wu, H., Xue, R., Tang, Z., Deng, C., Liu, T., Zeng, H., Sun, Y., & Shen, X. (2010). Metabolomic investigation of gastric cancer tissue using gas chromatography/mass spectrometry. Analytical and Bioanalytical Chemistry, 396(4), 1385–1395.
Ikeda, A., Nishiumi, S., Shinohara, M., Yoshie, T., Hatano, N., Okuno, T., Bamba, T., Fukusaki, E., Takenawa, T., Azuma, T., & Yoshida, M. (2012). Serum metabolomics as a novel diagnostic approach for gastrointestinal cancer. Biomedical Chromatography, 26(5), 548–558.
Song, H., Peng, J. S., Dong-Sheng, Y., Yang, Z. L., Liu, H. L., Zeng, Y. K., Shi, X. P., & Lu, B. Y. (2012). Serum metabolic profiling of human gastric cancer based on gas chromatography/mass spectrometry. Brazilian Journal of Medical and Biological Research, 45(1), 78–85.
Posada-Ayala, M., Zubiri, I., Martin-Lorenzo, M., Sanz-Maroto, A., Molero, D., Gonzalez-Calero, L., Fernandez-Fernandez, B., de la Cuesta, F., Laborde, C. M., Barderas, M. G., Ortiz, A., Vivanco, F., & Alvarez-Llamas, G. (2014). Identification of a urine metabolomic signature in patients with advanced-stage chronic kidney disease. Kidney International, 85, 103–111.
Acknowledgments
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant No. 81470196). The authors also thank BGI for the excellent technical assistance and are specifically grateful to Pro Aihua Zhang for many helpful discussions and suggestions.
Conflict of Interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 79 kb)
Rights and permissions
About this article
Cite this article
Liang, Q., Wang, C. & Li, B. Metabolomic Analysis Using Liquid Chromatography/Mass Spectrometry for Gastric Cancer. Appl Biochem Biotechnol 176, 2170–2184 (2015). https://doi.org/10.1007/s12010-015-1706-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1706-z