Abstract
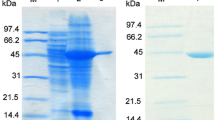
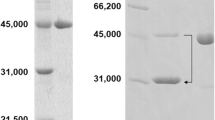
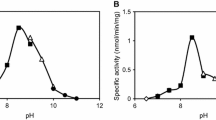
In silico identification for enzymes having desired functions is attractive because there is a possibility that numerous desirable enzymes have been deposited in databases. In this study, α-amino-ε-caprolactam (ACL) racemases were searched from the NCBI protein database. Four hundred thirteen fold-type I pyridoxal 5′-phosphate-dependent enzymes which are considered to contain sequences of ACL racemase were firstly obtained by submitting the sequence of ACL racemase from Achromobacter obae to the database. By identifying Lys241 as a key amino acid residue, 13 candidates for ACL racemase were selected. Then, putative ACL racemase genes were synthesized as codon-optimized sequences for expression in Escherichia coli. They were subcloned and expressed in E. coli BL21 and underwent His-tag purification. ACL and amino acid amide racemizing activities were detected among ten of the candidates. The locus tags Oant_4493, Smed_5339, and CSE45_2055 derived from Ochrobactrum anthropi ATCC49188, Sinorhizobium medicae WSM 419, and Citreicella sp. SE45, respectively, showed higher racemization activity against d- and l-ACLs rather than that of ACL racemase from A. obae. Our results demonstrate that the newly discovered ACL racemases were unique from ACL racemase from A. obae and might be useful for applications in dynamic kinetic resolution for d- or l-amino acid production.



Similar content being viewed by others
References
Gröger, H., & Asano, Y. (2012). Introduction-principles and historical landmarks of enzyme catalysis in organic synthesis. In K. Drauz, H. Gröger, & O. May (Eds.), Enzyme catalysis in organic synthesis (Vol. 2, pp. 3–42). Weinheim: Wiley-VCH Verlag & Co. KGaA.
Asano, Y. (2002). Overview of screening for new microbial catalysts and their uses in organic synthesis-selection and optimization of biocatalysts. Journal of Biotechnology, 94(1), 65–72.
Asano, Y. (2010). Tools for Enzyme Discovery -Industrial enzymes, biocatalysis and enzyme evolution. In H. B. Richard, E. D. Julian, & D. Arnold (Eds.), Manual of Industrial Microbiology and Biotechnology (pp. 441–452). USA: ASM press.
Bornscheuer, U. T., Huisman, G. W., Kazlauskas, R. J., Lutz, S., Moore, J. C., & Robins, K. (2012). Engineering the third wave of biocatalysis. Nature, 485, 185–194.
Gerlt, J. A., & Babbitt, P. C. (2000). Can sequence determine function? Genome Biology, 1(5), 0005.1–005.10.
Lee, D., Redfern, O., & Oregon, C. (2007). Prediction protein function from sequence and structure. Nature Reviews Molecular Cell Biology, 8, 995–1005.
Davids, T., Schmidt, M., Böttcher, D., & Bornscheuer, U. T. (2013). Strategies for the discovery and engineering of enzymes for biocatalysis. Current Opinion in Chemical Biology, 17, 215–220.
Höhne, M., Schätzle, S., Jochens, H., Robins, K., & Bornscheuer, U. T. (2010). Rational assignment of key motifs for function guides in silico enzyme identification. Nature Chemical Biology, 6, 807–813.
Loewenstein, Y., Raimondo, D., Redfern, O. C., Watson, J., Frishman, D., Linial, M., Orengo, C., Thornton, J., & Tramontano, A. (2009). Protein function annotation by homology-based inference. Genome Biology, 10(2), 207.1–207.8.
Steffen-Munsberg, F., Vickers, C., Kohls, H., Land, H., Mallin, H., Nobili, A., Skalden, L., Bergh, T. V. D., Joosten, H. J., Berglund, P., Höhne, M., & Bornscheuer, U. T. (2015). Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnology Advances. doi:10.1016/j.biotechadv.2014.12.012.
Fukumura, T. (1977). Bacterial racemization of α-amino-ε-caprolactam. Agricultural and Biology Chemistry, 41(8), 1321–1325.
Fukumura, T. (1977). Conversion of D- and DL-α-amino-ε-caprolactam into L-lysine using both yeast cells and bacterial cells. Agricultural and Biological Chemistry, 41(8), 1327–1330.
Fukumura, T. (1977). Partial purification and some properties of α-amino-ε-caprolactam-racemizing enzyme from Achromobacter obae. Agricultural and Biological Chemistry, 41(8), 1509–1510.
Ahmed, S. A., Esaki, N., Tanaka, H., & Soda, K. (1982). Production and stabilization of α-amino-ε-caprolactam racemase from Achromobacter obae. Bulletin of the Institute for Chemical Research, 60(5–6), 342–346.
Ahmed, S. A., Esaki, N., & Soda, K. (1982). Purification and properties of α-amino-ε-caprolactam racemase from Achromobacter obae. FEBS Letters, 150(2), 370–374.
Ahmed, S. A., Esaki, N., Tanaka, H., & Soda, K. (1983). Racemization of α-amino-δ-valerolactam catalyzed by α-amino-ε-caprolactam racemase from Achromobacter obae. Agricultural and Biological Chemistry, 47(5), 1149–1150.
Ahmed, S. A., Esaki, N., Tanaka, H., & Soda, K. (1983). Properties of α-amino-ε-caprolactam racemase from Achromobacter obae. Agricultural and Biological Chemistry, 47(8), 1887–1893.
Ahmed, S. A., Esaki, N., Tanaka, H., & Soda, K. (1984). L-α-Amino-β-thio-ε-caprolactam, a new sulfur-containing substrate for α-amino-ε-caprolactam racemase. FEBS Letters, 174(1), 76–79.
Ahmed, S. A., Esaki, N., Tanaka, H., & Soda, K. (1985). Mechanism of inactivation of α-amino-ε-caprolactam racemase by α-amino-δ-valerolactam. Agricultural and Biological Chemistry, 49(10), 2991–2997.
Fukumura, T. (1976). Screening, classification and distribution of L-α-amino-ε-caprolactam-hydrolysing yeasts. Agricultural and Biological Chemistry, 40(9), 1687–1693.
Fukumura, T. (1976). Hydrolysis of L-α-amino-ε-caprolactam by yeasts. Agricultural and Biological Chemistry, 40(9), 1695–1698.
Asano, Y., & Yamaguchi, S. (2005). Discovery of amino acid amides as new substrates for α-amino-ε-caprolactam racemase from Achromobacter obae. Journal of Molecular Catalysis B: Enzymatic, 36, 22–29.
Asano, Y., & Yamaguchi, S. (2005). Dynamic kinetic resolution of amino acid amide catalyzed by D-aminopeptidase and α-amino-ε-caprolactam racemase. Journal of the American Chemical Society, 127, 7696–7697.
Yamaguchi, S., Komeda, H., & Asano, Y. (2007). New enzymatic method of chiral amino acid synthesis by dynamic kinetic resolution of amino acid amides: use of stereoselective amino acid amidases in the presence of α-amino-ε-caprolactam racemase. Applied and Environmental Microbiology, 73(16), 5370–5373.
Okazaki, S., Suzuki, A., Mizushima, T., Kawano, T., Komeda, H., Asano, Y., & Yamane, T. (2009). The novel structure of a pyridoxal 5’-phosphate-dependent fold-type I racemase, α-amino-ε-caprolactam racemase from Achromobacter obae. Biochemistry, 48(5), 941–950.
Ahmed, S. A., Esaki, N., Tanaka, H., & Soda, K. (1986). Mechanism of α-amino-ε-caprolactam racemase reaction. Biochemistry, 25(2), 385–388.
Yoshimura, T., & Goto, M. (2008). D-amino acids in the brain: structure and function of pyridoxal phosphate-dependent amino acid racemases. FEBS Journal, 275(14), 3527–3537.
Nakano, S., & Asano, Y. (2015). Protein evolution analysis of S-hydroxynitrile lyase by complete sequence design utilizing the INTMSAlign software. Scientific Reports, 5, 1–10.
Pellegata, R., Pinza, M., & Pifferi, G. (1978). An improved synthesis of γ-, δ-, and ε-lactams. Synthesis, 8, 614–616.
Bradford, M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
McNicholas, S., Potterton, E., Wilson, K. S., & Noble, E. M. (2011). Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallographica Section D: Biological Crystallography, 67, 386–394.
Ito, T., Maekawa, M., Hayashi, S., Goto, M., Hemmi, H., & Yoshimura, T. (2013). Catalytic mechanism of serine racemase from Dictyostelium discoideum. Amino Acids, 44, 1073–1084.
Eliot, A. C., & Kirsch, J. F. (2004). Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annual Review of Biochemistry, 73, 383–415.
Boesten, W.H.J., Raemakers-Franken, P.C., Sonke, T., Euverink, G.J.W. & Grijpstra, P. (2003). Polypeptides having α-H-α-amino acid amide racemase activity and nucleic acids encoding the same. WO International Patent Application, WO2003106691.
Yasukawa, K., & Asano, Y. (2012). Enzymatic synthesis of chiral phenylalanine derivatives by a dynamic kinetic resolution of corresponding amide and nitrile substrates with a multi-enzyme system. Advanced Synthesis & Catalysis, 354(17), 3327–3332.
Maier, A., Riedlinger, J., Fiedler, H.-P., & Hampp, R. (2004). Actinomycetales bacteria from a spruce stand: characterization and effects on growth of root symbiotic and plant parasitic soil fungi in dual culture. Mycological Progress, 3(2), 129–136.
Acknowledgments
This work was financed by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (Grant No. PHD/099/2551) and Japan Student Services Organization (JASSO). The authors greatly appreciated Asst. Prof. Ken-ichi Fuhshuku for the preparation of the chiral ACLs and enantiomerically pure amino acid amides.
Conflict of Interest
None declared.
Ethical Approval
Not required.
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 423 kb)
Rights and permissions
About this article
Cite this article
Payoungkiattikun, W., Okazaki, S., Nakano, S. et al. In Silico Identification for α-Amino-ε-Caprolactam Racemases by Using Information on the Structure and Function Relationship. Appl Biochem Biotechnol 176, 1303–1314 (2015). https://doi.org/10.1007/s12010-015-1647-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1647-6