Abstract
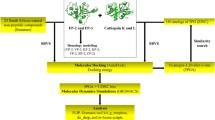
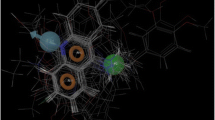
Malaria is one of the most infectious diseases in the world. Plasmodium vivax, the pathogen causing endemic malaria in humans worldwide, is responsible for extensive disease morbidity. Due to the emergence of resistance to common anti-malarial drugs, there is a continuous need to develop a new class of drugs for this pathogen. P. vivax cysteine protease, also known as vivapain-2, plays an important role in haemoglobin hydrolysis and is considered essential for the survival of the parasite. The three-dimensional (3D) structure of vivapain-2 is not predicted experimentally, so its structure is modelled by using comparative modelling approach and further validated by Qualitative Model Energy Analysis (QMEAN) and RAMPAGE tools. The potential binding site of selected vivapain-2 structure has been detected by grid-based function prediction method. Drug targets and their respective drugs similar to vivapain-2 have been identified using three publicly available databases: STITCH 3.1, DrugBank and Therapeutic Target Database (TTD). The second approach of this work focuses on docking study of selected drug E-64 against vivapain-2 protein. Docking reveals crucial information about key residues (Asn281, Cys283, Val396 and Asp398) that are responsible for holding the ligand in the active site. The similarity-search criterion is used for the preparation of our in-house database of drugs, obtained from filtering the drugs from the DrugBank database. A five-point 3D pharmacophore model is generated for the docked complex of vivapain-2 with E-64. This study of 3D pharmacophore-based virtual screening results in identifying three new drugs, amongst which one is approved and the other two are experimentally proved. The ADMET properties of these drugs are found to be in the desired range. These drugs with novel scaffolds may act as potent drugs for treating malaria caused by P. vivax.




Similar content being viewed by others
References
Carlton, J. M., et al. (2008). Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature, 455, 757–763.
Gardner, M. J., et al. (2002). Genome sequence of the human malaria parasite Plasmodium falciparum. Nature, 419, 498–511.
Sarduy, E. S., & Planes, M. d. l. A. C. (2013). Efficient expression systems for cysteine proteases of malaria parasites: too good to be true? Bioengineered, 4, 12–19.
Singh, N., et al. (2006). Plasmodium falciparum: biochemical characterization of the cysteine protease falcipain-2. Experimental Parasitology, 112, 187–192.
Na, B., et al. (2004). Identification and biochemical characterization of vivapains cysteine proteases of the malaria parasite Plasmodium vivax. Biochemistry Journal, 378, 529–538.
Benson, D. A., et al. (2013). GenBank. Nucleic Acids Research, 41, D36–D42.
Boratyn, G. M., et al. (2012). Domain enhanced lookup time accelerated BLAST. Biology Direct, 7, 12.
Notredame, C., et al. (2000). T-Coffee: a novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology, 302, 205–217.
Waterhouse, A. M., et al. (2009). Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics, 25, 1189–1191.
Eswar, N. et al. (2006), Comparative protein structure modeling using Modeller. Current Protocols in Bioinformatics, pp. 5.6.1–5.6.30.
Hildebrand, A., et al. (2009). Fast and accurate automatic structure prediction with HHpred. Proteins: Structure, Function, and Bioinformatics, 77, 128–132.
Guex, N., & Peitsch, M. C. (1997). SWISS‐MODEL and the Swiss‐Pdb Viewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723.
Fernandez-Fuentes, N., et al. (2007). M4T: a comparative protein structure modeling server. Nucleic Acids Research, 35, W363–W368.
Lindahl, E., et al. (2006). NOMAD-Ref: visualization. deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Research, 34, W52–W56.
Benkert, P., et al. (2009). QMEAN server for protein model quality estimation. Nucleic Acids Research, 37, W510–W514.
Lovell, S. C., et al. (2003). Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins, 50, 437–450.
Volkamer, A., et al. (2012). DoGSiteScorer: a web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics, 28, 2074–2075.
Kuhn, M., et al. (2012). STITCH 3: zooming in on protein–chemical interactions. Nucleic Acids Research, 40, D876–D880.
Knox, C., et al. (2011). DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Research, 39, D1035–D1041.
Zhu, F., et al. (2012). Therapeutic Target Database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Research, 40, D1128–D1136.
Morris, G. M., et al. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30, 2785–2791.
Wolber, G., & Langer, T. (2004). LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. Journal of Chemical Information and Modeling, 45, 160–169.
Kirchmair, J., et al. (2005). Comparative analysis of protein-bound ligand conformations with respect to catalyst's conformational space subsampling algorithms. Journal of Chemical Information and Modeling, 45, 422–430.
Molinspiration Chemoinformatics on the Web [Online]. Available: http://www.molinspiration.com.
Ghose, A. K., et al. (1999). A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery: 1. A qualitative and quantitative characterization of known drug databases. Journal of Combinatorial Chemistry, 1, 55–68.
Lipinski, C. A. (2004). Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technologies, 1, 337–341.
Lipinski, C. A., et al. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 46, 3–26.
Cheng, F., et al. (2012). admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. Journal of Chemical Information and Modeling, 52, 3099–3105.
Mason, C. S., et al. (2001). Baculoviral expression and characterization of rodent cathepsin S. Protein Expression and Purification, 23, 45–54.
White, A. C., Jr., et al. (1997). Characterization of a cysteine proteinase from Taenia crassiceps cysts. Molecular and icalBiochem Parasitology, 85, 243–253.
Semenov, A., et al. (1998). Antimalarial synergy of cysteine and aspartic protease inhibitors. Antimicrobial Agents and Chemotherapy, 42, 2254–2258.
Moon, S.-U., et al. (2011). Plasmodium vivax: collaborative roles for plasmepsin 4 and vivapains in hemoglobin hydrolysis. Experimental Prasitology, 128, 127–132.
Galewska, Z., & Bankowski, E. (1994). Cathepsin D—a main proteolytic enzyme of the bovine vitreous. Roczniki Akademii Medycznej w Bialymstoku, 39, 38–43.
Sahu, N. K., et al. (2008). Novel molecular targets for antimalarial drug development. Chemical Biology & Drug Design, 71, 287–297.
Jana, S., & Paliwal, J. (2007). Novel molecular targets for antimalarial chemotherapy. International Journal of Antimicrobial Agents, 30, 4–10.
Kuhn, D. J., et al. (2007). Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin–proteasome pathway, against preclinical models of multiple myeloma. Blood, 110, 3281–3290.
Acknowledgments
Manoj Kumar Yadav is grateful to the Indian Council of Medical Research (ICMR), India, for awarding him with the Senior Research fellowship. Amisha Singh is thankful to the Department of Bioinformatics, MMV, Banaras Hindu University, for providing computational facilities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yadav, M.K., Singh, A. & Swati, D. A Knowledge-Based Approach for Identification of Drugs Against Vivapain-2 Protein of Plasmodium vivax Through Pharmacophore-Based Virtual Screening with Comparative Modelling. Appl Biochem Biotechnol 173, 2174–2188 (2014). https://doi.org/10.1007/s12010-014-1023-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1023-y