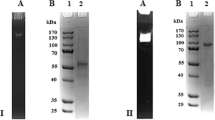
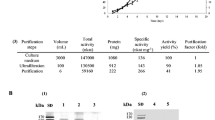
Abstract
Class I cellobiose dehydrogenases (CDHs) are extracellular hemoflavo enzymes produced at low levels by the Basidiomycetes (white rot fungi). In presence of suitable electron acceptors, e.g., cytochrome c, 2,6-dichlorophenol-indophenol, or metal ions, it oxidizes cellobiose to cellobionolactone. A stringent requirement for disaccharides makes CDH also useful for conversion of lactose to lactobionic acid, an important ingredient in pharma and detergent industry. In this work, class I CDH was produced using a newly identified white rot fungus Termitomyces sp. OE147. Four media were evaluated for CDH production, and maximum enzyme activity of 0.92 international unit (IU)/ml was obtained on Ludwig medium under submerged conditions. Statistical optimization of N source, which had significant effect on CDH production, using Box–Behnken design followed by optimization of inoculum size and age resulted in an increase in activity to 2.9 IU/ml and a productivity of ~25 IU/l/h. The nearly purified CDH exhibited high activity of 26.4 IU/mg protein on lactose indicating this enzyme to be useful for lactobionic acid synthesis. Some of the internal peptide sequences bore 100 % homology to the CDH produced in Myceliophthora thermophila. The fungal isolate was amenable to scale up, and an overall productivity of ~18 IU/l/h was obtained at 14-l level.





Similar content being viewed by others
References
Cameron, M. D., & Aust, S. D. (2001). Cellobiose dehydrogenase—an extracellular fungal flavocytochrome. Enzyme and Microbial Technology, 28, 129–138.
Zamocky, M., Ludwig, R., Peterbauer, C., Hallberg, B., Divne, C., & Nicholls, P. (2006). Cellobiose dehydrogenase—a flavocytochrome from wood—degrading, phytopathogenic and saprotropic fungi. Current Protein and Peptide Science, 7, 255–280.
Bao, W., Usha, S. N., & Renganathan, V. (1993). Purification and characterization of cellobiose dehydrogenase, a novel extracellular hemoflavoenzyme from the white-rot fungus Phanerochaete chrysosporium. Archives of Biochemistry and Biophysics, 300, 705–713.
Xu, F., Golightly, E. J., Duke, K. R., Lassen, S. F., Knusen, B., Christensen, S., et al. (2001). Humicola insolens cellobiose dehydrogenase: cloning, redox chemistry, and “logic gate”-like dual functionality. Enzyme and Microbial Technology, 28, 744–753.
Baminger, U., Subramaniam, S. S., Renganathan, V., & Haltrich, D. (2001). Purification and characterization of cellobiose dehydrogenase from the plant pathogen Sclerotium (Athelia) rolfsii. Applied and Environmental Microbiology, 67, 1766–1774.
Henriksson, G., Johansson, G., & Petterson, G. (2000). A critical review of cellobiose dehydrogenase. Journal of Biotechnology, 78, 93–113.
Henriksson, G., Zhang, L. J., Ljungquist, P., Reitberger, T., Petterson, G., & Johansson, G. (2000). Is cellobiose dehydrogenase from Phanerochaete chrysosporium a lignin degrading enzyme? Biochimica et Biophysica Acta, 1480, 83–91.
Dumonceaux, T., Bartholomew, K., Laleanu, L., Charles, T., & Archibald, F. (2001). Cellobiose dehydrogenase is essential for wood invasion and non-essential for Kraft pulp delignification by Trametes versicolor. Enzyme and Microbial Technology, 29, 478–489.
Dhariwal, A., Mavrov, V., & Scroeder, I. (2006). Production of lactobionic acid with process integrated electrochemical enzyme regeneration and optimisation of process variables using response surface methods (RSM). Journal of Molecular Catalysis B: Enzymatic, 42, 64–69.
Barbara, A., Green, R. P., Richard, H., Wildnauer, P. D., & Brenda, L. E. Lactobionic acid- a novel polyhydroxy bionic acid for skincare. Princeton: Neostrata Company, Inc.
Fang, J., Huang, F., & Gao, P. (1999). Optimization of cellobiose dehydrogenase production by Schizophyllum commune and effect of the enzyme on kraft pulp bleaching by ligninases. Process Biochemistry, 34, 957–961.
Harreither, W., Sygmund, C., Dunhofen, E., Vicuna, R., Haltrich, D., & Ludwig, R. (2009). Celloboise dehydrogenase from the ligninolytic basidiomycetes Ceriporiopsis subvermispora. Applied and Environmental Microbiology, 75, 2750–2757.
Li, B., Rotsaert, A. J. F., Gold, M. H., & Renganathan, V. (2000). Homologous expression of recombinant cellobiose dehydrogenase in Phanerochaete chrysosporium. Biochemical and Biophysical Research Communications, 270, 141–146.
Yoshida, M., Ohira, T., Igarashi, K., Nagasawa, H., Aida, K., Hallberg, B. M., et al. (2001). Production and characterization of recombinant Phanerochaete chrysosporium cellobiose dehydrogenase in the methylotrophic yeast Pichia pastoris. Bioscience, Biotechnology, and Biochemistry, 65, 2050–2057.
Stapleton, P. C., O’Brien, M. M., O’Callaghan, J., & Dobson, A. D. W. (2004). Molecular cloning of the cellobiose dehydrogenase from Trametes versicolor and expression in Pichia pastoris. Enzyme and Microbial Technology, 34, 55–63.
Zamocky, M., Schümann, C., Sygmund, C., Dobson, A. D., Ludwig, R., Haltrich, D., et al. (2008). Cloning, sequence analysis and heterologous expression in Pichia pastoris of a gene encoding a thermostable cellobiose dehydrogenase from Myriococcum thermophilum. Protein Expression and Purification, 59, 258–265.
Zhang, R., Fan, Z., & Kasuga, T. (2011). Expression of cellobiose dehydrogenase from Neurospora crassa in Pichia pastoris and its purification and characterization. Protein Expression and Purification, 75, 63–69.
Ludwig, R., Salamon, A., Varga, J., Zamocky, M., Peterbauer, C. K., & Kulbe, K. D. (2004). Characterization of cellobiose dehydrogenases from the white-rot fungi Trametes pubescens and Trametes villosa. Applied Microbiology and Biotechnology, 64, 213–222.
Connick, D., Bouquelet, J., & Dumoitiers, D. (2000). Industrial media and fermentation processes for improved growth and protease production by Tetrahymena thermophila. Journal of Industrial Microbiology and Biotechnology, 24, 285–290.
Saha, T., Ghosh, D., Mukherjee, S., Bose, S., & Mukherjee, M. (2008). Cellobiose dehydrogenase production by the mycelial culture of the mushroom Termitomyces clypeatus. Process Biochemistry, 48, 634–641.
Nystrom, J. M., & DiLuca, P. H. Enhanced production of Trichoderma cellulase on high levels of cellulose in submerged culture. IIT Delhi Symposium 1977.
Biswas, R., Sahai, V., Mishra, S., & Bisaria, V. S. (2010). Bioprocess strategies for enhanced production of xylanase by Melanocarpus albomyces IITD3A on agro—residual extract. Journal of Bioscience and Bioengineering, 110, 702–708.
Box, G. E. P., & Behnken, D. W. (1960). Some new three level design for the study of quantitative variables. Technometrics, 2, 455–475.
Baminger, U., Nidetzky, B., Kulbe, K. D., & Haltrich, D. (1999). A simple assay for measuring cellobiose dehydrogenase activity in the presence of laccase. Journal of Microbiological Methods, 35, 253–259.
Canevascini, G., Borer, P., & Dreyer, J. L. (1991). Cellobiose dehydrogenase of Sporotricum (Chrysosporium) thermophile. European Journal of Biochemistry, 198, 43–52.
Ghosh, T. K. (1987). Measurement of cellulase activities. Pure and Applied Chemistry, 59, 257–268.
Rogalski, J., Szczodrak, J., & Janusz, G. (2006). Manganese peroxidase production in submerged cultures by free and immobilized mycelia of Nematoloma frowardii. Bioresource Technology, 97, 469–476.
Arora, D. S., & Gill, P. K. (2001). Comparison of two assay procedures for lignin peroxidase. Enzyme and Microbial Technology, 28, 602–605.
Temp, U., & Eggert, C. (1999). Novel interaction between laccase and cellobiose dehydrogenase during pigment synthesis in the white rot fungus Pycnoporus cinnabarinus. Applied and Environmental Microbiology, 65, 389–395.
Meyer, S. P., & Ahearn, D. G. (1977). Extracellular proteolysis by Candida lipolytica. Mycologia, 69, 646–651.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry, 72, 248–254.
Song, A. R., Tian, X. M., Feng, G. F., & Zhang, Y. Q. (2001). A study of the utilization of Pleurotus eryngii with different carbon and nitrogen sources. Acta Edulis Fungi, 8, 10–14.
Qinnghe, C., Xiaoyu, Y., Tiangui, N., Cheng, J., & Qiugang, M. (2004). The screening of culture conditions and properties of xylanase by white-rot fungus Pleurotus ostreatus. Process Biochemistry, 39, 1561–1566.
Gruno, M., Valjamae, P., Pettersson, G., & Johansson, G. (2004). Inhibition of the Trichoderma ressei cellulases by cellobiose is strongly dependent on the nature of the substrate. Biotechnology and Bioengineering, 86, 503–511.
Subramaniam, S. S., Nagalla, S. R., & Reganathan, V. (1999). Cloning and characterization of a thermostable cellobiose dehydrogenase from Sporotrichum thermophile. Archives of Biochemistry and Biophysics, 365, 223–230.
Yoshida, M., Ohira, T., Igarashi, K., Nagasawa, H., & Samejima, M. (2002). Molecular cloning and characterization of a cDNA encoding cellobiose dehydrogenase from the wood-rotting fungus Grifola frondosa. FEMS Microbiology Letters, 217, 225–230.
Bailey, J. E., & Ollis, D. F. (1986) Biochemical engineering fundamentals, 2nd Ed. New York: McGraw Hill, Inc.
Sikyta, B. (1983). Development of microbial processes. In B. Sikyta & M. Prause (Eds.), Methods in industrial microbiology (pp. 250–274). Chichester: Ellis Horwood Ltd.
Archibald, F. S., Bourbonnais, R., Jurasek, L., Paice, M. G., & Reid, I. D. (1997). Kraft pulp bleaching and delignification by Trametes versicolor. Journal of Biotechnology, 53, 215–236.
Kudriavtseva, O. A., Dunaevskii, L. E., Kamzolkina, O. V., & Belozerskii, M. A. (2008). Proteolytic enzymes of the fungi: extracellular proteases of xylotrophic basidiomycetes. Mikrobiologiia, 77, 725–737.
Sabotic, J., Trcek, T., Popovic, T., & Brzin, J. (2007). Basidiomycets harbor a hidden treasure of proteolytic activity. Journal of Biotechnology, 128, 297–307.
Acknowledgments
Authors gratefully acknowledge the financial assistance received from the Department of Biotechnology (Government of India) for carrying out this study. Further, we are thankful to Dr. R. C. Upadhyay from the National Research Centre for Mushroom (NRCM) Solan, India, for providing the cultures. The authors would also like to acknowledge the help of Mr. M.V.R.K. Sarma for the analysis of the statistical data.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
(DOC 52 kb)
Supplementary Fig. 2
(DOC 37 kb)
Rights and permissions
About this article
Cite this article
Gupta, G., Gangwar, R., Gautam, A. et al. Production of Cellobiose Dehydrogenase from a Newly Isolated White Rot Fungus Termitomyces sp. OE147. Appl Biochem Biotechnol 173, 2099–2115 (2014). https://doi.org/10.1007/s12010-014-1010-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1010-3