Abstract
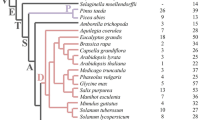
Plant proteome databases were mined for a flavin monooxygenase (YUCCA), tryptophan decarboxylase (TDC), nitrilase (NIT), and aldehyde oxidase (AO) enzymes that could be involved in the tryptophan-dependent pathway of auxin biosynthesis. Phylogenetic trees for enzyme sequences obtained were constructed. The YUCCA and TDC trees showed that these enzymes were conserved across the plant kingdom and therefore could be involved in auxin synthesis. YUCCAs branched into two clades. Most experimentally studied YUCCAs were found in the first clade. The second clade which has representatives from only seed plants contained Arabidopsis sequences linked to embryonic development. Therefore, sequences in this clade were suggested to be evolved with seed development. Examination of TDC activity and expression had previously linked this enzyme to secondary products synthesis. However, the phylogenetic finding of a conserved TDC clade across land plants suggested its essential role in plant growth. Phylogenetic analysis of AOs showed that plants inherited one AO. Recent gene duplication was suggested as AO sequences from each species were similar to each other rather than to AO from other species. Taken together and based on the experimental support of the involvement of AO in abscisic synthesis, AO was excluded as an intermediate in IAA production. Phylogenetic tree for NIT showed that the first clade contained sequences from species across the plant kingdom whereas the second branch contained sequences from only Brassicaceae. Even though NIT4 orthologues were conserved in the second clade, their major role seems to be detoxification of hydrogen cyanide rather than producing IAA.







Similar content being viewed by others
References
Stepanova, A., et al. (2008). Cell, 133(1), 177–191.
Tao, Y., et al. (2008). Cell, 133(1), 164–176.
Yamada, M., et al. (2009). Plant Physiology, 151, 168–179.
Nafisi, M., et al. (2007). Plant Cell, 19(6), 2039–2052.
Normanly, J., et al. (1997). Plant Cell, 9(10), 1781–1790.
Bak, S., et al. (2001). Plant Cell, 13(1), 101–111.
Noe, W., Mollenschott, C., & Berlin, J. (1984). Plant Molecular Biology, 3(5), 281–288.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). Nucleic Acids Research, 22, 4673–4680.
Fitch, W. M. (1981). Journal of Molecular Evolution, 18(1), 30–37.
Fitch, W. M. (1971). Systematic Zoology, 20, 406–416.
Felsenstein, J. (1981). Journal of Molecular Evolution, 17(6), 368–376.
Dayhoff, M. O. (1978), in A model of evolutionary change in proteins, vol. 5: Atlas of Protein Sequence and Structure (Dayhoff, M.O., ed.), Washington, D.C., pp 345–352.
Kimura, M. (1983) The neutral theory of molecular evolution, Cambridge, pp 50–56.
George, D. G., Hunt, L. T., & Barker, W. C. (1988), in Current methods in sequence comparison and analysi, vol. 2: Macromolecular Sequencing and Synthesis (Schlesinger, D.H. and Liss, A.R., ed.), New York, pp 127–149.
Felsenstein, J. (1989). Cladistics, 5, 164–166.
Zhao, Y. D., et al. (2001). Science, 291(5502), 306–309.
Exposito-Rodriguez, M., et al. (2007). Journal of Plant Growth Regulation, 26(4), 329–340.
Gallavotti, A., et al. (2008). Proceedings of the National Academy of Sciences of the United States of America, 105(39), 15196–15201.
Tobena-Santamaria, R., et al. (2002). Genetics Development, 16(6), 753–763.
Yamamoto, Y., et al. (2007). Plant Physiology, 143, 1362–1371.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Nucleic Acids Research, 25, 3389–3402.
Yamazaki, Y., et al. (2003). Plant & Cell Physiology, 44(4), 395–403.
LopezMeyer, M., & Nessler, C. L. (1997). Plant Journal, 11(6), 1167–1175.
De Luca, V., Marineau, C., & Brisson, N. (1989). Proceedings of the National Academy of Sciences of the United States of America, 86(8), 2582–2586.
Seo, M., et al. (2000). Plant Journal, 23(4), 481–488.
Rajagopal, R. (1971). Physiology Plantarum, 24, 272–281.
Bower, P. J., Brown, H. M., & Purves, W. K. (1978). Plant Physiology, 61(1), 107–110.
Koshiba, T., et al. (1996). Plant Physiology, 110(3), 781–789.
Park, W. J., et al. (2003). Plant Physiology, 133(2), 794–802.
ANGIS/BioManager. 2009, The University of Sydney.
Page, R. D. M. (1996). Computer Applications in the Biosciences, 12, 357–358.
Woo, Y. M., et al. (2007). Plant Molecular Biology, 65(1–2), 125–136.
Kaminaga, Y., et al. (2006). Journal of Biological Chemistry, 281(33), 23357–23366.
Kawalleck, P., et al. (1993). Journal of Biological Chemistry, 268(3), 2189–2194.
Cheng, Y. F., Dai, X. H., & Zhao, Y. D. (2006). Genetics Development, 20(13), 1790–1799.
Cheng, Y. F., Dai, X. H., & Zhao, Y. D. (2007). Plant Cell, 19(8), 2430–2439.
Kang, S., et al. (2008). Planta, 227(1), 263–272.
Facchini, P. J., & De Luca, V. (1995). Phytochemistry, 38(5), 1119–1126.
Marques, I. A., & Brodelius, P. E. (1988). Plant Physiology, 88(1), 52–55.
Islas, I., Loyolavargas, V. M., & Mirandaham, M. D. (1994). In Vitro Cell Dev-Pl, 30(1), 81–83.
Songstad, D. D., et al. (1990). Plant Physiology, 94(3), 1410–1413.
Zdunek-Zastocka, E., et al. (2004). Journal of Experimental Botany, 55(401), 1361–1369.
Gonzalez-Guzman, M., et al. (2004). Plant Physiology, 135(1), 325–333.
Seo, M., et al. (2000). Proceedings of the National Academy of Sciences of the United States of America, 97(23), 12908–12913.
Vorwerk, S., et al. (2001). Planta, 212(4), 508–516.
Bartel, B., & Fink, G. (1994). Proceedings of the National Academy of Sciences, 91(14), 6649–6653.
Bartling, D., et al. (1994). Proceedings of the National Academy of Sciences, 91(13), 6021–6025.
Wittstock, U., & Halkier, B. (2002). Trends in Plant Science, 7(6), 263–270.
Piotrowski, M., Schonfelder, S., & Weiler, E. W. (2001). Journal of Biological Chemistry, 276(4), 2616–2621.
Jenrich, R., et al. (2007). Proceedings of the National Academy of Sciences of the United States of America, 104(47), 18848–18853.
Kriechbaumer, V., et al. (2007). Journal of Experimental Botany, 58(15–16), 4225–4233.
Pollmann, S., Düchting, P., & Weiler, E. (2009). Phytochemistry, 70(4), 523–531.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abu-Zaitoon, Y.M. Phylogenetic Analysis of Putative Genes Involved in the Tryptophan-Dependent Pathway of Auxin Biosynthesis in Rice. Appl Biochem Biotechnol 172, 2480–2495 (2014). https://doi.org/10.1007/s12010-013-0710-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0710-4