Abstract
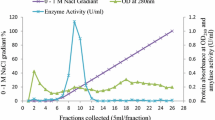
A novel α-amylase (McAmyA) from the thermophilic fungus, Malbranchea cinnamomea was purified, characterized and crystallized in the present study. McAmyA was purified to apparent homogeneity with a molecular mass of 60.3 kDa on SDS-PAGE. The enzyme exhibited maximal activity at pH 6.5 and was stable within pH 5.0–10.0. It was most active at 65 °C and was stable up to 50 °C. McAmyA was capable of hydrolyzing amylose, starch, amylopectin, pullulan, cyclodextrins and maltooligosaccharides. The full-length cDNA of an α-amylase gene (McAmyA) from the strain was cloned. McAmyA consisted of a 1,476-bp open reading frame encoding 492 amino acids. It displayed the highest amino acid sequence homology (less than 60 %) with the reported α-amylases. The crystal structure of McAmyA was solved at a resolution of 2.25 Å (PDB code 3VM7). The overall structure of McAmyA reveals three domains with ten α helices and 14 β strands, and the putative catalytic residues are positioned at domain A with somewhat different secondary structural circumstances compared with typical α-amylases.




Similar content being viewed by others
Abbreviations
- CAPS:
-
(Cyclohexylamino)-1-propanesulphonic acid
- CD:
-
Cyclodextrin
- CHES:
-
2-(cyclohexylamino) ethanesulfonic acid
- GH:
-
Glycoside hydrolase
- McAmyA:
-
An α-amylase from M. cinnamomea S168
- MES:
-
2-(N-morpholino)ethane sulfonic acid
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- RACE:
-
Rapid amplification of cDNA ends
- TLC:
-
Thin-layer chromatography
References
Derde, L. J., Gomand, S. V., Courtin, C. M., & Delcour, J. A. (2012). Characterisation of three starch degrading enzymes: Thermostable β-amylase, maltotetraogenic and maltogenic α-amylases. Food Chemistry, 135, 713–721.
Gupta, R., Gigras, P., Mohapatra, H., Goswami, V. K., & Chauhan, B. (2003). Microbial α-amylases: a biotechnological perspective. Process Biochemistry, 38, 1599–1616.
Maarel, M. J., Veen, B., Uitdehag, J. C., Leemhuis, H., & Dijkhuizen, L. (2002). Properties and applications of starch-converting enzymes of the α-amylase family. Journal of Biotechnology, 94, 137–155.
Prakash, O., & Jaiswal, N. (2010). α-Amylase: An ideal representative of thermostable enzymes. Applied Microbiology and Biotechnology, 160, 2401–2414.
Bai, Y., Huang, H., Meng, K., Shi, P., Yang, P., Luo, H., et al. (2012). Identification of an acidic α-amylase from Alicyclobacillus sp. A4 and assessment of its application in the starch industry. Food Chemistry, 131, 1473–1478.
Kumar, S., & Khare, S. K. (2012). Purification and characterization of maltooligosaccharide-forming α-amylase from moderately halophilic Marinobacter sp. EMB8. Bioresource Technology, 116, 247–251.
Aquino, A. C. M. M., Jorge, J. A., Terenzi, H. F., & Polizeli, M. L. T. M. (2003). Studies on a thermostable α-amylase from the thermophilic fungus Scytalidium thermophilum. Applied Microbiology and Biotechnology, 61, 323–328.
Hernandez, M. S., Rodrıguez, M. R., Guerra, N. P., & Roses, R. P. (2006). Amylase production by Aspergillus niger in submerged cultivation on two wastes from food industries. Journal of Food Engineering, 73, 93–100.
Michelin, M., Silva, T. M., Benassi, V. M., Peixoto-Nogueira, S. C., Moraes, L. A. B., Leão, J. M., et al. (2010). Purification and characterization of a thermostable α-amylase produced by the fungus Paecilomyces variotii. Carbohydrate Research, 345, 2348–2353.
Li, S., Zuo, Z., Niu, D., Singh, S., Permaul, K., Prior, B. A., et al. (2011). Gene cloning, heterologous expression, and characterization of a high maltose-producing α-amylase of Rhizopus oryzae. Applied Biochemistry and Biotechnology, 164, 581–592.
Petrova, S. D., Ilieva, S. Z., Bakalova, N. G., Atev, A. P., Bhat, M. K., & Kolev, D. N. (2000). Production and characterization of extracellular α-amylases from the thermophilic fungus Thermomyces lanuginosus (wild and mutant strains). Biotechnology Letters, 22, 1619–1624.
Matsuura, Y., Kusunoki, M., Harada, W., & Kakudo, M. (1984). Structure and possible catalytic residues of Taka-amylase A. Journal of Biochemistry, 95, 697–702.
Boel, E., Brady, L., Brzozowski, A. M., Derewenda, Z., Dodson, G. G., Jensen, V. J., et al. (1990). alcium binding in α-amylases: an x-ray diffraction study at 2.1-Å resolution of two enzymes from Aspergillus. Biochemistry, 29, 6244–6249.
Gautam, S. P., & Gupta, A. K. (1992). Extraeellular and mycelial amylases of the thermophilie fungus Malbranchea sulfurea. Mycopathologia, 119, 77–82.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 277, 680–685.
Leatherbarrow, R. J. (1999). GraFit Version 4.0.10. UK: Erithacus Software Limited.
Sambrook, J., & Russell, D. W. (2001). Molecular cloning: a laboratory manual (3rd ed.). New York: Cold Spring Harbor.
Rose, T. M., Henikoff, J. G., & Henikoff, S. (2003). CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Research, 31, 3763–3766.
Evans, P. (2005). Scaling and assessment of data quality. Acta Crystallographica Section D: Biological Crystallography, 62, 72–82.
Winn, M. D., Bollard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallographica Section D: Biological Crystallography, 67, 235–242.
Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L. W., et al. (2008). Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallographica Section D: Biological Crystallography, 64, 61–69.
Emsley, P., & Gowtan, K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallographica Section D: Biological Crystallography, 60, 2126–2132.
Yao, M., Zhou, Y., & Tanaka, I. (2006). Software for automating the refinement process of protein structure analysis. Acta Crystallographica Section D: Biological Crystallography, 62, 189–196.
Vagin, A. A., Steiner, R. A., Lebedev, A. A., Potterton, L., McNicholas, S., Long, F., et al. (2004). REFMACS dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallographica Section D: Biological Crystallography, 60, 2184–2195.
Oshoma, C. E., Imarhiagbe, E. E., Ikenebomeh, M. J., & Eigbaredon, H. E. (2010). Nitrogen supplements effect on amylase production by Aspergillus niger using cassava whey medium. African Journal of Biotechnology, 9, 682–686.
Champreda, V., Kanokratana, P., Sriprang, R., Tanapongpipat, S., & Eurwilaichitrp, L. (2007). Purification, biochemical characterization, and gene cloning of a new extracellular thermotolerant and glucose tolerant maltooligosaccharide-forming α-amylase from an endophytic ascomycete Fusicoccum sp. BCC4124. Bioscience, Biotechnology, and Biochemistry, 71, 2010–2020.
Hostinová, E., Janeček, S., & Gašperik, J. (2010). Gene sequence, bioinformatics and enzymatic characterization of α-amylase from Saccharomycopsis fibuligera KZ. The Protein Journal, 29, 355–364.
Pandey, A., Nigam, P., Soccol, C. R., Soccol, V., Singh, D., & Mohan, R. (2000). Advances in microbial amylases. Biotechnology and Applied Biochemistry, 31, 135–152.
Seong-Ae, Y., Ryu, S., Lee, S., & Moon, T. (2008). Purification and characterization of branching specificity of a novel extracellular amylolytic enzyme from marine hyperthermophilic Rhodothermus marinus. Journal of Microbiology and Biotechnology, 18, 457–464.
Wanderley, K. J., Torres, F. A. G., Moraes, L. M. P., & Ulhoa, C. J. (2004). Biochemical characterization of α-amylase from the yeast Crytococcus flavus. FEMS Microbiology Letters, 231, 165–169.
Dheeran, P., Kumar, S., Jaiswal, Y. K., & Adhikari, D. K. (2010). Characterization of hyperthermostable α-amylase from Geobacillus sp. IIPTN. Applied Microbiology and Biotechnology, 86, 1857–1866.
Kim, I. C., Cha, J. H., Kim, J. R., Jang, S. Y., Seo, B. C., Cheong, T. K., et al. (1992). Catalytic properties of the cloned amylase from Bacillus licheniformis. The Journal of Biological Chemistry, 267, 22108–22114.
Park, K. H., Kim, T. J., Cheong, T. K., Kim, J. W., Oh, B. H., & Svensson, B. (2000). Structure, specificity and function of cyclomaltodextrinase, a multispecific enzyme of the α-amylase family. Biochimica et Biophysica Acta, 1478, 165–185.
Kato, S., Shimizo-Ibika, A., Mura, K., Takeuchi, A., Tokue, C., & Arai, S. (2007). Molecular cloning and characterization of an α-amylase from Pichia burtonii 15–1. Bioscience, Biotechnology, and Biochemistry, 71, 3007–3013.
Shimura, Y., Oh, K., Kon, M., Yamamoto, E., Mizuno, Y., Adachi, T., et al. (2011). Enzymatic synthesis of novel branched sugar alcohols mediated by the transglycosylation reaction of pullulan-hydrolyzing amylase II (TVA II) cloned from Thermoactinomyces vulgaris R-47. Carbohydrate Research, 346, 1842–1847.
Tonozuka, T., Mogi, S., Shimura, Y., Ibuka, A., Sakai, H., Matsuzawa, H., et al. (1995). Comparison of primary structures and substrate specificities of two pullulan-hydrolyzing α-amylases, TVA I and TVA II, from Thermoactinomyces vulgaris R-47. Biochimica et Biophysica Acta, 1252, 35–42.
Holm, L., & Rosenstrom, P. (2010). Dali server: conservation mapping in 3D. Nucleic Acids Research, 38, 545–549.
Zhou, Y., Yao, M., & Tanaka, I. (2006). New algorithm for protein model building: extending a partial model in a map segment. Journal of Applied Crystallography, 39, 57–63.
Tanaka, A., & Hoshino, E. (2003). Secondary calcium-binding parameter of Bacillus amyloliquefaciens α-amylase obtained from inhibition kinetics. Journal of Bioscience and Bioengineering, 96, 262–267.
Acknowledgments
This work was supported by a research grant from the National High Technology Research and Development Program of China (863 Program, no. 2011AA100905).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Han, P., Zhou, P., Hu, S. et al. A Novel Multifunctional α-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure. Appl Biochem Biotechnol 170, 420–435 (2013). https://doi.org/10.1007/s12010-013-0198-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0198-y