Abstract
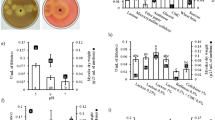
A high cellobiohydrolase (CBH)-producing strain was isolated and identified as Penicillium purpurogenum KJS506 according to the morphology and comparison of internal transcribed spacer rDNA gene sequence. When rice straw and corn steep powder were used as carbon and nitrogen sources, respectively, a maximum CBH activity of 2.6 U mg-protein−1, one of the highest among CBH-producing microorganisms, was obtained. The optimum temperature and pH for CBH production were 30 °C and 4.0, respectively. The increased production of CBH in P. purpurogenum culture at 30 °C was confirmed by two-dimensional electrophoresis followed by MS/MS sequencing of the partial peptide. The internal amino acid sequences of P. purpurogenum CBH showed a significant homology with hydrolases from glycoside hydrolase family 7. The extracellular CBH was purified to homogeneity by sequential chromatography of P. purpurogenum culture supernatants on a DEAE-sepharose column, a gel filtration column, and then on a Mono Q column with fast-protein liquid chromatography. The purified CBH was a monomeric protein with a molecular weight of 60 kDa and showed broad substrate specificity with maximum activity towards p-nitrophenyl β-d-cellobiopyranoside. P. purpurogenum CBH showed t 1/2 value of 4 h at 60 °C and V max value of 11.9 μmol min−1 mg-protein−1 for p-nitrophenyl-d-cellobiopyranoside. Although CBHs have been reported, the high specific activity distinguishes P. purpurogenum CBH.





Similar content being viewed by others
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
Baldrian, P., & Valaskova, V. (2008). Degradation of cellulose by Basidiomycetous fungi. FEMS-Microbiology Reviews, 32, 501–521.
Bhat, M. K., & Bhat, S. (1997). Cellulose degrading enzymes and their potential industrial applications. Biotechnology Advances, 15, 583–620.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Eriksson, K., Blanchette, R. A., & Ander, P. (1990). Microbial and enzymatic degradation of wood and wood components. Berlin: Springer.
Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochemical Journal, 280, 309–316.
Hong, J., Tamaki, H., Yamamoto, K., & Kumagai, H. (2003). Cloning of a gene encoding thermostable cellobiohydrolase from Thermoascus aurantiacus and its expression in yeast. Applied Microbiology and Biotechnology, 63, 42–50.
Hou, Y., Wang, T., Long, H., & Zhu, H. (2007). Cloning, sequencing and expression analysis of the first cellulase gene encoding cellobiohydrolase 1 from a cold-adaptive Penicillium chrysogenum FS010. Acta Biochim Biophys Sin (Shanghai), 39, 101–107.
Igarashi, K., Samejima, M., & Eriksson, K. E. (1998). Cellobiose dehydrogenase enhances Phanerochaete chrysosporium cellobiohydrolase I activity by relieving product inhibition. European Journal of Biochemistry, 253, 101–106.
Kanokratana, P., Chantasingh, D., Champreda, V., Tanapongpipat, S., Pootanakit, K., & Eurwilaichitr, L. (2008). Identification and expression of cellobiohydrolase (CBHI) gene from an endophytic fungus, Fusicoccum sp. (BCC4124) in Pichia pastoris. Protein Expression and Purification, 58, 148–153.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Lahjouji, K., Storms, R., Xiao, Z., Joung, K. B., Zheng, Y., Powlowski, J., et al. (2007). Biochemical and molecular characterization of a cellobiohydrolase from Trametes versicolor. Applied Microbiology and Biotechnology, 75, 337–346.
Li, Y. L., Li, D. C., & Teng, F. C. (2006). Purification and characterization of a cellobiohydrolase from the thermophilic fungus Chaetomium thermophilus CT2. Wei Sheng Wu Xue Bao, 46, 143–6.
Limam, F., Chaabouni, S., Ghrir, R., & Marzouki, N. (1995). Two cellobiohydrolases of Penicillium occitanis mutant Pol 6: Purification and properties. Enz Microbial Technol, 17, 340–346.
Lynd, L. R., Laser, M. S., Bransby, D., Dale, B. E., Davison, B., Hamilton, R., et al. (2008). How biotech can transform biofuels. Nature Biotechnology, 26, 169–172.
Mathew, G. M., Sukumaran, R. K., Singhania, R. R., & Pandey, A. (2008). Progress in research on fungal cellulases for lignocellulose degradation. Journal of Scientific and Industrial Research, 67, 898–907.
Medve, J., Lee, D., & Tjerneld, F. (1998). Ion-exchange chromatographic purification and quantitative analysis of Trichoderma reesei cellulases cellobiohydrolase I, II and endoglucanase II by fast protein liquid chromatography. J Chromatography A, 808, 153–165.
Miller, G. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analy Chem, 31, 426–428.
O’Farrell, P. H. (1975). High resolution two-dimensional electrophoresis of proteins. Journal of Biological Chemistry, 250, 4007–4021.
Ohnishi, Y., Nagase, M., Ichiyanagi, T., Kitamoto, Y., & Aimi, T. (2007). Transcriptional regulation of two cellobiohydrolase encoding genes (cel1 and cel2) from the wood-degrading Basidiomycete Polyporus arcularius. Applied Microbiology and Biotechnology, 76, 1069–1078.
Parkkinen, T., Koivula, A., Vehmaanpera, J., & Rouvinen, J. (2008). Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding. Protein Science, 17, 1383–1394.
Rouau, X., & Odier, E. (1986). Purification and properties of two enzymes from Dichomitus squalens which exhibit both cellobiohydrolase and xylanase activity. Carbohydrate Research, 145, 279–292.
Shevchenko, A., Wilm, M., Vorm, O., & Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry, 68, 850–858.
Shoemaker, S., Schweickart, V., Ladner, M., Gelfand, D., Kwok, S., Myambo, K., et al. (1983). Molecular cloning of exo–cellobiohydrolase I derived from Trichoderma reesei strain l27. Bio/Technology, 1, 691–696.
Stahl, P., & Klug, M. (1996). Characterization and differentiation of filamentous fungi based on fatty acid composition. Applied and Environmental Microbiology, 62, 4136–4146.
Stahlberg, J., Divne, C., Koivula, A., Piens, K., Claeyssens, M., Teeri, T. T., et al. (1996). Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei. Journal of Molecular Biology, 264, 337–349.
Sun, X., Liu, Z., Qu, Y., & Li, X. (2008). The effects of wheat bran composition on the production of biomass-hydrolyzing enzymes by Penicillium decumbens. Applied Biochemistry and Biotechnology, 146, 119–128.
Takao, S., Kamagata, Y., & Sasaki, H. (1985). Cellulase production by Penicillium purpurogenum. Journal of Fermentation Technology, 63, 127–134.
Teeri, T. T., Koivula, A., Linder, M., Wohlfahrt, G., Divne, C., & Jones, T. A. (1998). Trichoderma reesei cellobiohydrolase: Why so efficient on crystalline cellulose? Biochemical Society Transactions, 26, 173–178.
Teeri, T. T., Lehtovaara, P., Kauppinen, S., Salovuori, I., & Knowles, J. (1987). Homologous domains in Trichoderma reesei cellulolytic enzymes: Gene sequence and expression of cellobiohydrolase II. Gene, 51, 43–52.
Tomme, P., Kwan, E., Gilkes, N. R., Kilburn, D. G., & Warren, R. A. (1996). Characterization of CenC, an enzyme from Cellulomonas fimi with both endo- and exoglucanase activities. Journal of Bacteriology, 178, 4216–4223.
Tomme, P., Van Tilbeurgh, H., Pettersson, G., Van Damme, J., Vandekerckhove, J., Knowles, J., et al. (1988). Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. European Journal of Biochemistry, 170, 575–581.
Tuohy, M. G., Walsh, D. J., Murray, P. G., Claeyssens, M., Cuffe, M. M., Savage, A. V., et al. (2002). Kinetic parameters and mode of action of the cellobiohydrolases produced by Talaromyces emersonii. Biochimica Et Biophysica Acta, 1596, 366–380.
Uzcategui, E., Ruiz, A., Montesino, R., Johansson, G., & Pettersson, G. (1991). The 1, 4-beta-D-glucan cellobiohydrolases from Phanerochaete chrysosporium. I. A system of synergistically acting enzymes homologous to Trichoderma reesei. Journal of Biotechnology, 19, 271–285.
Wang, T., Wang, C., Gao, P., Zhong, L., & Zou, Y. (1998). Subcloning and expression of coding region for cellulase binding domain of CBH I from P. janthinellum in E. coli. Wei Sheng Wu Xue Bao, 38, 269–75.
White, T., Bruns, T., & Lee, S. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. Innis & T. White (Eds.), PCR protocols: A guide to methods and applications (pp. 315–322). New York: Academic.
Zhang, Y. H. P. (2009). A sweet out-of-the-box solution to the hydrogen economy: Is the sugar-powered car science fiction? Energy Environ Sci, 2, 272–282.
Zhang, Y. H. P., Himmel, M. E., & Mielenz, J. R. (2006). Outlook for cellulase improvement: Screening and selection strategies. Biotechnology Advances, 24, 452–481.
Acknowledgments
This study was supported by a grant (code 2008A0080126) from ARPC. It was also supported by a grant (Code 20070301034024) from BioGreen 21 Program, Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, KM., Joo, AR., Jeya, M. et al. Production and Characterization of Cellobiohydrolase from a Novel Strain of Penicillium purpurogenum KJS506. Appl Biochem Biotechnol 163, 25–39 (2011). https://doi.org/10.1007/s12010-010-9013-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-010-9013-1