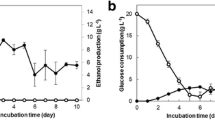
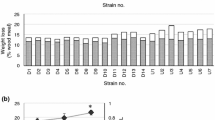
Abstract
Ligninolytic enzymes are well-known to play the crucial roles in lignin biodegradation and have potential applications in industrial processes. The filamentous white-rot fungus, Phanerochaete chrysosporium, has been widely used as a model organism for studying these ligninolytic enzymes that are able to degrade the lignin during the secondary metabolism. To study the gene expression in secondary metabolism and metabolic switching phase of P. chrysosporium, we constructed a metabolic-switching phase suppression subtractive hybridization (SSH) cDNA library and a secondary metabolic phase SSH cDNA library to compare their mRNA expression profiles. We isolated the genes that are specially expressed and subsequently identified four genes that specially expressed during metabolic-switching phase while 22 genes in secondary metabolic phase. Accordingly, these specially expressed genes might play key roles in different metabolic stages, which would offer more new insights into the shift from nitrogen to lignin metabolism.





Similar content being viewed by others
References
Adler, E. (1977). Lignin chemistry—past, present and future. Wood Science and Technology, 11, 169–218.
Kiem, R., & Kögel-Knabner, I. (2003). Contribution of lignin and polysaccharides to the refractory carbon pool in C-depleted arable soils. Soil Biology and Biochemistry, 35(1), 101–118.
Kersten, P., & Cullen, D. (2007). Extracellular oxidative systems of the lignin-degrading Basidiomycete Phanerochaete chrysosporium. Fungal Genetics and Biology, 44(2), 77–87.
Reddy, C. A., & D'Souza, T. M. (1994). Physiology and molecular biology of the lignin peroxidases of Phanerochaete chrysosporium. FEMS Microbiology Reviews, 13(2–3), 137–152.
Buswell, J. A., Odier, E., & Kent, K. T. (1987). Lignin biodegradation. Critical Reviews in Biotechnology, 6, 1–60.
Garg, S. K., & Modi, D. R. (1999). Decolorization of pulp-paper mill effluents by white-rot fungi. Critical Reviews in Biotechnology, 19(2), 85–112.
Wesenberg, D., Kyriakides, I., & Agathos, S. N. (2003). White-rot fungi and their enzymes for the treatment of industrial dye effluents. Biotechnology Advances, 22(1–2), 161–187.
Martinez, D., Larrondo, L. F., Putnam, N., Gelpke, M. D., Huang, K., Chapman, J., et al. (2004). Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nature Biotechnology, 22(6), 695–700.
Vanden Wymelenberg, A., Minges, P., Sabat, G., Martinez, D., Aerts, A., Salamov, A., et al. (2006). Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveal complex mixtures of secreted proteins. Fungal Genetics and Biology, 43(5), 343–356.
Keyser, P., Kirk, T. K., & Zeikus, J. G. (1978). Ligninolytic enzyme system of Phanaerochaete chrysosporium synthesized in the absence of lignin in response to nitrogen starvation. Journal of Bacteriology, 135(3), 790–797.
Boominathan, K., D'Souza, T. M., Naidu, P. S., Dosoretz, C., & Reddy, C. A. (1993). Temporal expression of the major lignin peroxidase genes of Phanerochaete chrysosporium. Applied and Environmental Microbiology, 59(11), 3496–3950.
Gold, M., & Alic, M. (1993). Molecular biology of the lignindegrading basidiomycete Phanerochaete chrysosporium. Microbiology and Molecular Biology Reviews, 57(3), 605–622.
Matsuzaki, F., Shimizu, M., & Wariishi, H. (2008). Proteomic and metabolomic analyses of the white-rot fungus Phanerochaete chrysosporium exposed to exogenous benzoic acid. Journal of Proteome Research, 7(6), 2342–2350.
Masahiko, M., Kureha, O., Mori, M., Kamitsuji, H., Suzuki, K., & Irie, T. (2007). Long serial analysis of gene expression for transcriptome profiling during the initiation of ligninolytic enzymes production in Phanerochaete chrysosporium. Applied Microbiology and Biotechnology, 75(3), 609–618.
Semarjit, S., Kapich, A. N., Panisko, E. A., Magnuson, J. K., Cullen, D., & Hammel, K. E. (2008). Differential expression in Phanerochaete chrysosporium of membrane-associated proteins relevant to lignin degradation. Applied and Environmental Microbiology, 74(23), 7252–7257.
Ozcan, S., Yildirim, V., Kaya, L., Albrecht, D., Becher, D., Hecker, M., et al. (2007). Phanerochaete chrysosporium soluble proteome as a prelude for the analysis of heavy metal stress response. Proteomics, 7(8), 1249–1260.
Vanden Wymelenberg, A., Gaskell, J., Mozuch, M., Kersten, P., Sabat, G., Martinez, D., et al. (2009). Transcriptome and secretome analysis of phanerochaete chrysosporium reveal complex patterns of gene expression. Applied and Environmental Microbiology, 75(12), 4058–4068.
Wymelenberg, A. V., Sabat, G., Martinez, D., Rajangam, A. S., Teeri, T. T., Gaskell, J., et al. (2005). The Phanerochaete chrysosporium secretome: database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. Journal of Biotechnology, 118(1), 17–34.
Jiang, M., Li, X., Zhang, L., Feng, H., & Zhang, Y. (2009). Gene expression analysis of Phanerochaete chrysosporium during the transition time from primary growth to secondary metabolism. Journal of Microbiology, 47(3), 308–318.
Zhang, Y. Z., Zylstra, G. J., Olsen, R. H., & Reddy, C. A. (1986). Identification of cDNA clones for ligninase from using synthetic oligonucleotide probes. Biochemical and Biophysical Research Communications, 137(2), 649–656.
Tien, M., & Kirk, T. K. (1988). Lignin peroxidase of Phanerochaete chrysosporium. Methods in Enzymology, 161, 238–349.
Kanehisa, M., Goto, S., Furumichi, M., Tanabe, M., & Hirakawa, M. (2009). KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Research. doi:10.1093/nar/gkp896.
Cullen, D. (1997). Recent advances on the molecular genetics of ligninolytic fungi. Journal of Biotechnology, 53(2–3), 273–289.
Diatchenko, L., Lau, Y. F., Campbell, A. P., Chenchik, A., Moqadam, F., Huang, B., et al. (1996). Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Science, 93(12), 6025–6030.
Diatchenko, L., Lukyanov, S., Lau, Y. F., & Siebert, P. D. (1999). Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods in Enzymology, 303, 349–380.
Finazzi Agrò, A., Federici, G., Giovagnoli, C., Cannella, C., & Cavallini, D. (1972). Effect of sulfur binding on rhodanese fluorescence. European Journal of Biochemistry, 28(1), 83–93.
Westley, J. (1973). Rhodanese. Advances in Enzymology and Related Areas of Molecular Biology, 39, 327–368.
Dhirendra, L. N., Paul, M. H., & John, W. (2000). Rhodanese as a thioredoxin oxidase. The International Journal of Biochemistry & Cell Biology, 32(4), 465–473.
Godon, C., Lagniel, G., Lee, J., Buhler, J. M., Kieffer, S., Perrot, M., et al. (1998). The H2O2 Stimulon in Saccharomyces cerevisiae. The Journal of Biological Chemistry, 273(35), 22480–22489.
Nordberg, J., & Arnér, E. S. (2001). Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radical Biology & Medicine, 31(11), 1287–1312.
Scharf, C., Riethdorf, S., Ernst, H., Engelmann, S., Völker, U., & Hecker, M. (1998). Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. Journal of Bacteriology, 180(7), 1869–1877.
Shi, M. M., Iwamoto, T., & Forman, H. J. (1994). gamma-Glutamylcysteine synthetase and GSH increase in quinone-induced oxidative stress in BPAEC. American Journal of Physiology. Lung Cellular and Molecular Physiology, 67(4), L414–L421.
Grant, C. M., Perrone, G., & Dawes, I. W. (1998). Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochemical and Biophysical Research Communications, 253(3), 893–898.
Grant, C. M., MacIver, F. H., & Dawes, I. W. (1997). Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to an accumulation of the dipeptide gamma-glutamylcysteine. Molecular Biology of the Cell, 8(9), 1699–1707.
Mehdi, K., & Penninckx, M. J. (1997). An important role for glutathione and {gamma}-glutamyltranspeptidase in the supply of growth requirements during nitrogen starvation of the yeast Saccharomyces cerevisiae. Microbiology, 143(6), 1885–1889.
Lee, J., Godon, C., Lagniel, G., Spector, D., Garin, J., Labarre, J., et al. (1999). Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. The Journal of Biological Chemistry, 274(23), 16040–16046.
Daniel, G., Volc, J., Filonova, L., Plihal, E., Kubatova, E., & Halada, P. (2007). Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Applied and Environmental Microbiology, 73, 6241–6253.
Assmann, E. M., Ottoboni, L. M., Ferraz, A., Rodríguez, J., & De Mello, M. P. (2003). Iron-responsive genes of Phanerochaete chrysosporium isolated by differential display reverse transcription polymerase chain reaction. Environmental Microbiology, 5(9), 777–786.
Miura, D., Tanaka, H., & Wariishi, H. (2004). Metabolic differential display analysis of the white-rot basidiomycete Phanerochaete chrysosporium grown under air and 100% oxygen. FEMS Microbiology Letters, 234(1), 111–116.
Lucic, E., Fourrey, C., Kohler, A., Martin, F., Chalot, M., & Brun-Jacob, A. (2008). A gene repertoire for nitrogen transporters in Laccaria bicolor. The New Phytologist, 180(2), 343–364.
Benjdia, M., Rikirsch, E., Müller, T., Morel, M., Corratgé, C., Zimmermann, S., et al. (2006). Peptide uptake in the ectomycorrhizal fungus Hebeloma cylindrosporum: characterization of two tripeptide transporters (HcPTR2A and B). The New Phytologist, 170(2), 401–410.
Larrondo, L. F., Vicuña, R., & Cullen, D. (2005). Phanerochaete chysosporium genomics. Applied Mycology and Biotechnology, 5, 315–352.
Acknowledgements
We thank Dr. Hong Feng, Chuan He, and Fan Bai for their critical reviews on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, JM., Zhang, Yz. Gene Expression in Secondary Metabolism and Metabolic Switching Phase of Phanerochaete chrysosporium . Appl Biochem Biotechnol 162, 1961–1977 (2010). https://doi.org/10.1007/s12010-010-8973-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-010-8973-5