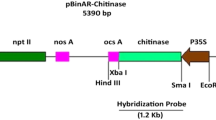
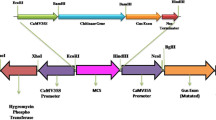
Abstract
The chit1 gene from the entomopathogenic fungus Metarhizium anisopliae, encoding the endochitinase CHIT42, was placed under the control of the CaMV 35S promoter, and the resulting construct was transferred to tobacco. Seventeen kanamycin-resistant transgenic lines were recovered, and the presence of the transgene was confirmed by polymerase chain reactions and Southern blot hybridization. The number of chit1 copies was determined to be varying from one to four. Copy number had observable effects neither on plant growth nor development. Substantial heterogeneity concerning production of the recombinant chitinase, and both general and specific chitinolytic activities were detected in leaf extracts from primary transformants. The highest chitinase activities were found in plants harboring two copies of chit1 inserts at different loci. Progeny derived from self-pollination of the primary transgenics revealed a stable inheritance pattern, with transgene segregation following a mendelian dihybrid ratio. Two selected plants expressing high levels of CHIT42 were consistently resistant to the soilborne pathogen Rhizoctonia solani, suggesting a direct relationship between enzyme activity and reduction of foliar area affected by fungal lesions. To date, this is the first report of resistance to fungal attack in plants mediated by a recombinant chitinase from an entomopathogenic and acaricide fungus.





Similar content being viewed by others
References
Duo-Chuan, L. (2006). Review of fungal chitinases. Mycopathologia, 161, 345–360. doi:10.1007/s11046-006-0024-y.
Dana, M. M., Pintor-Toro, J. A., & Cubero, B. (2006). Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiology, 142, 722–730. doi:10.1104/pp.106.086140.
Lorito, M., Harman, G. E., Hayes, C. K., Broadway, R. M., Tronsmo, A., Woo, S. L., et al. (1993). Chitinolytic enzymes of Trichoderma harzianum—purification of chitobiosidase and endochitinase. Phytopathology, 83, 302–307. doi:10.1094/Phyto-83-302.
Lorito, M., Hayes, C. K., Di Pietro, A., Woo, S. L., & Harman, G. E. (1994). Purification, characterisation and synergistic activity of a glucan 1,3-beta-glucosidase and an N-acetyl-glucosaminidase from Trichoderma harzianum. Phytopathology, 84, 398–405. doi:10.1094/Phyto-84-398.
Lorito, M., Mach, R. L., Sposato, P., Strauss, J., Peterbauer, C. K., & Kubicek, C. P. (1996). Mycoparasitic interaction relieves binding of the Cre1 carbon catabolite repressor protein to promoter sequences of the ech42 (endochitinase-encoding) gene in Trichoderma harzianum. Proceedings of the National Academy of Sciences of the United States of America, 93, 14868–14872. doi:10.1073/pnas.93.25.14868.
Seidl, V., Huemer, B., Seiboth, B., & Kubicek, C. P. (2005). A complete survey of Trichoderma chitinases reveals three distinct subgroups of family 18 chitinases. FEBS Journal, 272, 5923–5939. doi:10.1111/j.1742-4658.2005.04994.x.
Dahiya, N., Tewari, R., & Hoondal, G. S. (2006). Biotechnological aspects of chitinolytic enzymes: a review. Applied Microbiology and Biotechnology, 71, 773–782. doi:10.1007/s00253-005-0183-7.
Lorito, M., Woo, S. L., Fernandez, I. G., Colucci, G., Harman, G. E., Pintor-Toro, J. A., et al. (1998). Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proceedings of the National Academy of Sciences of the United States of America, 95, 7860–7865. doi:10.1073/pnas.95.14.7860.
Gokul, B., Lee, J. H., Song, K. B., Rhee, S. K., Kim, C. H., & Panda, T. (2000). Characterization and applications of chitinases from Trichoderma harzianum—a review. Bioprocess Engineering, 23, 691–694. doi:10.1007/s004499900138.
Bolar, J. P., Norelli, J. L., Wong, K. W., Hayes, C. K., Harman, G. E., & Aldwinckle, H. S. (2000). Expression of endochitinase from Trichoderma harzianum in transgenic apple increases resistance to scab and reduces vigour. Phytopathology, 90, 72–77. doi:10.1094/PHYTO.2000.90.1.72.
Bolar, J. P., Norelli, J. L., Harman, G. E., Brown, S. K., & Aldwinckle, H. S. (2001). Synergistic activity of endochitinase and exochitinase from Trichoderma atroviride (T. harzianum) against the pathogenic fungus (Venturia inaequalis) in transgenic apple plants. Transgenic Research, 10, 533–543. doi:10.1023/A:1013036732691.
Noël, A., Levasseur, C., Le, V. Q., & Seguin, A. (2005). Enhanced resistance to fungal pathogens in forest trees by genetic transformation of black spruce and hybrid poplar with a Trichoderma harzianum endochitinase gene. Physiological and Molecular Plant Pathology, 67, 92–99. doi:10.1016/j.pmpp.2005.09.010.
Emani, C., Garcia, J. M., Lopata-Finch, E., Pozo, M. J., Uribe, P., Kim, D. J., et al. (2003). Enhanced fungal resistance in transgenic cotton expressing an endochitinase gene from Trichoderma virens. Plant Biotechnology Journal, 1, 321–336. doi:10.1046/j.1467-7652.2003.00029.x.
Terakawa, T., Takaya, N., Horiuchi, H., Koike, M., & Takagi, M. (1997). A fungal chitinase gene from Rhizopus oligosporus confers antifungal activity to transgenic tobacco. Plant Cell Reports, 16, 439–443.
St. Leger, R. J., Cooper, R. M., & Charnley, A. K. (1991). Characterization of a chitinase and chitobiase produced by the entomopathogenic fungus Metarhizium anisopliae. Journal of Invertebrate Pathology, 58, 415–426. doi:10.1016/0022-2011(91)90188-V.
Pinto, A. S., Barreto, C. C., Schrank, A., Ulhoa, C. J., & Vainstein, M. H. (1997). Purification and characterization of an extracellular chitinase from the entomopathogen Metarhizium anisopliae. Canadian Journal of Microbiology, 43, 322–327.
Bogo, M. R., Rota, C. A., Pinto, H., Jr., Ocampos, M., Correa, C. T., Vainstein, M. H., et al. (1998). A chitinase encoding gene (chit1 gene) from the entomopathogen Metarhizium anisopliae: isolation and characterisation of genomic and full-length cDNA. Current Microbiology, 73, 221–225. doi:10.1007/s002849900368.
Kang, S. C., Park, S., & Lee, D. G. (1999). Purification and characterization of a novel chitinase from the entomopathogenic fungus Metarhizium anisopliae. Journal of Invertebrate Pathology, 73, 276–281. doi:10.1006/jipa.1999.4843.
Frazzon, A. P., Vaz Junior, I., Masuda, A., Schrank, A., & Vainstein, M. H. (2000). In vitro assessment of Metarhizium anisopliae isolates to control the cattle tick Boophilus microplus. Veterinary Parasitology, 94, 117–125. doi:10.1016/S0304-4017(00)00368-X.
da Silva, M. V., Santi, L., Staats, C. C., da Costa, A. M., Colodel, E. M., Driemeier, D., et al. (2005). Cuticle-induced endo/exoacting chitinase CHIT30 from Metarhizium anisopliae is encoded by an ortholog of the chi3 gene. Research in Microbiology, 156, 382–392. doi:10.1016/j.resmic.2004.10.013.
St. Leger, R. J., Charnley, K., & Cooper, R. M. K. (1986). Cuticle degrading enzymes of entopathogenic fungi: mechanisms of interaction between pathogen enzymes and insect cuticle. Journal of Invertebrate Pathology, 47, 117–125.
St. Leger, R. J., Joshi, L., Bidochka, M., Rizzos, M. J., & Roberts, D. W. (1996). Characterisation and ultrastructural location of chitinases from Metarhizium anisopliae, M. flavoviridae and Beauveria bassiana during fungal invasion of host (Manduca sexta) cuticle. Applied and Environmental Microbiology, 62, 907–912.
Sambrook, J., & Russel, D. W. (2001). Molecular cloning: a laboratory manual (3rd ed.). New York: Cold Spring Harbor Laboratory Press.
Peach, C. R. W., & Velten, J. (1994). Agrobacterium-mediated gene transfer to plant cells: cointegrate and binary vector systems. In S. B. Gelvin, R. A. Schilperoort & D. P. S. Verma (Eds.), Plant molecular biology manual, Section B1 (pp. 1–19). Dordrecht: Kluwer.
Horsch, R. B., Fry, J. E., Hoffmann, N. I., Eichholtz, D., Rogers, S. G., & Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science, 277, 1229–1231.
Doyle, J. J., & Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissues. Phytochemical Bulletin, 19, 11–150.
Armitage, P., & Berry, G. (1987). Statistical methods in medical research (2nd ed.). London: Oxford University Press.
Memelink, J., Swords, K. M. M., Staehelin, L. A., & Hoge, J. H. C. (1994). Southern, northern and western blot analysis. In S. B. Gelvin, R. A. Schilperoort & D. P. S. Verma (Eds.), Plant molecular biology manual, Section F1 (pp. 1–23). Dordrecht: Kluwer.
Jefferson, R. A., & Wilson, K. J. (1994). The gus gene fusion system. In S. B. Gelvin, R. A. Schilperoort & D. P. S. Verma (Eds.), Plant molecular biology manual, Section B14 (pp. 9–33). Dordrecht: Kluwer.
Bradford, M. M. (1976). A rapid and sensitive method for quantitation of microgram quantities of protein utilising the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254. doi:10.1016/0003-2697(76)90527-3.
Reissig, J. L., Stromnger, L., & Leloir, L. F. (1955). A modified colorimetric method for the determination of N-acetylamino sugars. Journal of Biological Chemistry, 217, 959–966.
Yabuki, M., Mizushima, K., Amatou, T., Ando, A., Fuji, I., Shimada, M., et al. (1986). Purification and characterisation of chitinase and a chitobiase produced by Aeromonas hydrophila subesp anaraegenes A42. The Journal of General and Applied Microbiology, 32, 25–32. doi:10.2323/jgam.32.25.
St. Leger, R. J., Staples, R. C., & Roberts, D. W. (1993). Entomopathogenic isolates of Metarhizium anisopliae, Beauveria bassiana and Aspergillus flavus produce multiple extracellular chitinase isozymes. Journal of Invertebrate Pathology, 61, 81–84. doi:10.1006/jipa.1993.1014.
Murashige, T., & Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiologia Plantarum, 15, 473–497. doi:10.1111/j.1399-3054.1962.tb08052.x.
Rohini, V. K., & Rao, K. S. (2001). Transformation of peanut (Arachis hypogaea L.) with tobacco chitinase gene: variable response of transformants to leaf spot disease. Plant Science, 160, 889–898. doi:10.1016/S0168-9452(00)00462-3.
Kishimoto, K., Nishizawa, Y., Tabei, Y., Hibi, T., Nakajima, M., & Akutsu, K. (2002). Detailed analysis of rice chitinase gene expression in transgenic cucumber plants showing different levels of disease resistance to grey mold (Botrytis cinerea). Plant Science, 162, 655–662. doi:10.1016/S0168-9452(01)00602-1.
Chai, B., Maqbool, S. B., Hajela, R. K., Green, D., Vargas, J. M., Jr., Warkentin, D., et al. (2002). Cloning of a chitinase-like cDNA (hs2), its transfer to creeping bentgrass (Agrostis palustris Huds.) and development of brown patch (Rhizoctonia solani) disease resistant transgenic lines. Plant Science, 163, 183–193. doi:10.1016/S0168-9452(02)00069-9.
Finnegan, J., & McElroy, D. (1994). Transgene inactivation: plants fight back!. Biotechnologies, 12, 883–888. doi:10.1038/nbt0994-883.
Maqbool, S. B., & Christou, P. (1999). Multiple traits of agronomic importance in transgenic indica rice plants: analysis of transgene integration patterns, expression levels and stability. Molecular Breeding, 5, 471–480. doi:10.1023/A:1009634226797.
Gooday, G. W., Zhu, W. Y., & Donnell, R. W. (1992). What are the roles of chitinases in the growing fungus? FEMS Microbiology Letters, 100, 387–392. doi:10.1111/j.1574-6968.1992.tb05730.x.
Datta, K., Jumin, T., Oliva, N., Ona, I., Velazhahan Mew, T. W., Muthukrishnan, S., et al. (2001). Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Science, 160, 405–414. doi:10.1016/S0168-9452(00)00413-1.
Deineko, E. V., Novoselya, T. V., Zagorskaya, A. A., Filipenko, E. A., & Shumnyi, V. K. (2000). Expression instability of the marker nptII gene in transgenic tobacco plants. Russian Journal of Plant Physiology: A Comprehensive Russian Journal on Modern Phytophysiology, 47, 394–399.
Acknowledgements
We thank Dr. A.T.S. Matsumura for providing the isolates of R. solani employed in this study, Dr. M.H. Bodanese-Zanettini for green house facilities, Dr. S.C. Jacques for her assistance with statistical analysis, and architect R. Pasquali for his help with the illustrations. This research was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/Brazil) and Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kern, M.F., Maraschin, S.d.F., Vom Endt, D. et al. Expression of a Chitinase Gene from Metarhizium anisopliae in Tobacco Plants Confers Resistance against Rhizoctonia solani . Appl Biochem Biotechnol 160, 1933–1946 (2010). https://doi.org/10.1007/s12010-009-8701-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-009-8701-1