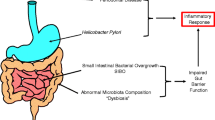
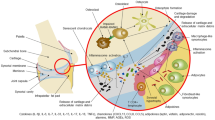
Abstract
Purpose of Review
The purpose of this review is to highlight recent evidence with respect to expression and metabolomic profiling in axial spondyloarthritis (axSpA) that included ankylosing spondylitis (AS).
Recent Findings
AxSpA is not only characterized by the strongest genetic contribution for any complex rheumatic disease but is also influenced by environmental and immunological factors. Large-scale association-based studies have identified over 100 genetic variants contributing to 30% of the genetic risk of ankylosing spondylitis. Recent studies in global expression and metabolomic profiling appear to highlight common themes despite differences in tissues, populations, techniques, and relative paucity of patients in many of these studies.
Summary
Expression studies support a role for immunomodulation and bone remodeling in the pathogenesis and progression of axSpA/AS, while metabolomic studies implicate the importance of the intestinal microbial metabolism as well as fat and choline metabolic pathways in AS.

Similar content being viewed by others
Abbreviations
- A1:
-
Adenosine A1 receptor
- A2AAR:
-
Adenosine A2A receptor
- A2BAR:
-
Adenosine A2B receptor
- ANTXR2:
-
Anthrax toxin receptor 2
- AS:
-
Ankylosing spondylitis
- ATG16L1:
-
Autophagy related 16 like 1
- axSpA:
-
Axial spondyloarthritis
- ASAS:
-
Assessment of spondyloarthritis international society
- ASDAS:
-
Ankylosing spondylitis disease activity score
- BASDAI:
-
Bath ankylosing spondylitis disease activity index
- BASFI:
-
Bath ankylosing spondylitis functional index
- BMD:
-
Bone mineral density
- BMP:
-
Bone morphogenetic protein
- BMP-2:
-
Bone morphogenetic protein 2
- BMP-4:
-
Bone morphogenetic protein 4
- BMP-7:
-
Bone morphogenetic protein 7
- BXDC5:
-
Brix domain-containing protein 5
- CRP:
-
C-reactive protein
- CTX:
-
Carboxy-terminal collagen crosslinks
- DKK1:
-
Dickkopf-related protein 1
- DKK3:
-
Dickkopf-related protein 3
- DEGs:
-
Differentially expressed genes
- EGFR:
-
Epidermal growth factor receptor
- ERA:
-
Enthesitis-related arthritis
- ESR:
-
Erythrocyte sedimentation rate
- GC-MS:
-
Gas chromatography-mass spectrometry
- GO:
-
Gene ontology
- GSK3β:
-
Glycogen synthase kinase 3 beta
- GWAS:
-
Genome-wide association studies
- HLA-B:
-
Human leukocyte antigen B
- HSP90AA1:
-
Heat shock protein 90 alpha family class A member 1
- IDO:
-
Indoleamine 2,3-dioxygenase
- IFN:
-
Interferon
- Ihh:
-
Indian hedgehog
- IL-1:
-
Interleukin-1
- IL-1β:
-
Interleukin-1 beta
- IL2RA:
-
Interleukin-2 receptor alpha
- IL2RB:
-
Interleukin-2 receptor beta
- IL-6:
-
Interleukin-6
- IL-17A:
-
Interleukin-17A
- IL-22:
-
Interleukin-22
- IL-23:
-
Interleukin-23
- IL-23R:
-
Interleukin-23 receptor
- IRGM:
-
Immunity related GTPase M
- lncRNA:
-
Long non-coding RNA
- ITM2A:
-
Integral membrane protein 2A
- JAK:
-
Janus kinase
- JIA:
-
Juvenile idiopathic arthritis
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- KIR3DL2:
-
Killer cell immunoglobulin-like receptor 3DL2
- KREMEN1:
-
Kremen protein 1
- LC-MS:
-
Liquid chromatography-mass spectrometry
- MAP3K7:
-
Mitogen-activated protein kinase kinase kinase 7
- MHC:
-
Major histocompatibility complex
- MGP:
-
Matrix gla protein
- MIF:
-
Macrophage migration inhibitory factor
- miRNA:
-
Micro-ribonucleic acid
- MMP-2:
-
Matrix metalloproteinase-2
- mRNA:
-
Messenger ribonucleic acid
- MMP-1:
-
Matrix metalloproteinase-1
- MMP-3:
-
Matrix metalloproteinase-3
- MS:
-
Mass spectrometry
- mSASSS:
-
Modified stoke ankylosing spondylitis spinal score
- NFKB1:
-
Nuclear factor kappa-B p105 subunit
- NMR:
-
Nuclear magnetic resonance
- NR4A2:
-
Nuclear receptor subfamily 4 group A member 2
- OA:
-
Osteoarthritis
- OPG:
-
Osteoprotegerin
- OPLS-DA:
-
Orthogonal projection to latent structure discriminant analysis
- PADI4:
-
Peptidyl arginine deiminase 4
- PBMCs:
-
Peripheral blood mononuclear cells
- PCA:
-
Principal component analysis
- PD-1:
-
Programmed cell death protein 1
- PDCD4:
-
Programmed cell death 4
- PLS-DA:
-
Partial least squares discriminant analysis
- PTGER4:
-
Prostaglandin E2 receptor 4
- RA:
-
Rheumatic arthritis
- RNAseq:
-
Ribonucleic acid sequencing
- SMAD5:
-
SMAD family member 5
- SMAD7:
-
SMAD family member 7
- SNP:
-
Single nucleotide polymorphism
- STAT:
-
Signal transducer and activator of transcription
- STAT1:
-
Signal transducer and activator of transcription 1
- STAT4:
-
Signal transducer and activator of transcription 4
- TG:
-
Triglycerides
- Tim-4:
-
T-cell immunoglobulin and mucin domain containing 4
- TLR4:
-
Toll-like receptor 4
- TLR5:
-
Toll-like receptor 5
- TNF:
-
Tumor necrosis factor
- TNF-α:
-
Tumor necrosis factor-alpha
- TNFAIP:
-
Tumor necrosis factor, alpha-induced protein 3
- TNFSF10:
-
TNF superfamily member 10
- ucMGP:
-
Uncarboxylated matrix gla protein
- VEGF:
-
Vascular endothelial growth factor A
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
• Hanson A, Brown MA. Genetics and the causes of ankylosing spondylitis. Rheum Dis Clin N Am. 2017;43(3):401–14. https://doi.org/10.1016/j.rdc.2017.04.006. State of the art review on the current status of genetic studies in Ankylosing Spondylitis.
Li Z, Brown MA. Progress of genome-wide association studies of ankylosing spondylitis. Clin Transl Immunol. 2017;6(12):e163. https://doi.org/10.1038/cti.2017.49.
Pimentel-Santos FM, Ligeiro D, Matos M, Mourao AF, Costa J, Santos H, et al. Whole blood transcriptional profiling in ankylosing spondylitis identifies novel candidate genes that might contribute to the inflammatory and tissue-destructive disease aspects. Arthritis Res Ther. 2011;13(2):R57. https://doi.org/10.1186/ar3309.
Assassi S, Reveille JD, Arnett FC, Weisman MH, Ward MM, Agarwal SK, et al. Whole-blood gene expression profiling in ankylosing spondylitis shows upregulation of toll-like receptor 4 and 5. J Rheumatol. 2011;38(1):87–98. https://doi.org/10.3899/jrheum.100469.
Ma H, Xu D, Fu Q. Identification of ankylosing spondylitis-associated genes by expression profiling. Int J Mol Med. 2012;30(3):693–6. https://doi.org/10.3892/ijmm.2012.1047.
• Fang F, Pan J, Xu L, Li G, Wang J. Identification of potential transcriptomic markers in developing ankylosing spondylitis: a meta-analysis of gene expression profiles. Biomed Res Int. 2015;2015:826316. https://doi.org/10.1155/2015/826316. First meta-analysis of gene expression profiling in AS.
Duan R, Leo P, Bradbury L, Brown MA, Thomas G. Gene expression profiling reveals a downregulation in immune-associated genes in patients with AS. Ann Rheum Dis. 2010;69(9):1724–9. https://doi.org/10.1136/ard.2009.111690.
Thomas GP, Duan R, Pettit AR, Weedon H, Kaur S, Smith M, et al. Expression profiling in spondyloarthropathy synovial biopsies highlights changes in expression of inflammatory genes in conjunction with tissue remodelling genes. BMC Musculoskelet Disord. 2013;14:354. https://doi.org/10.1186/1471-2474-14-354.
Xu L, Sun Q, Jiang S, Li J, He C, Xu W. Changes in gene expression profiles of the hip joint ligament of patients with ankylosing spondylitis revealed by DNA chip. Clin Rheumatol. 2012;31(10):1479–91. https://doi.org/10.1007/s10067-012-2038-9.
•• Zhang C, Wang C, Jia Z, Tong W, Liu D, He C, et al. Differentially expressed mRNAs, lncRNAs, and miRNAs with associated co-expression and ceRNA networks in ankylosing spondylitis. Oncotarget. 2017;8(69):113543–57. https://doi.org/10.18632/oncotarget.22708. This study was the first to combine mRNAs, lncRNAs, and microRNAs to investigate AS pathogenesis.
Chen K, Wei XZ, Zhu XD, Bai YS, Chen Y, Wang CF, et al. Whole-blood gene expression profiling in ankylosing spondylitis identifies novel candidate genes that may contribute to the inflammatory and tissue-destructive disease aspects. Cell Immunol. 2013;286(1–2):59–64. https://doi.org/10.1016/j.cellimm.2013.10.009.
Lv Q, Li Q, Zhang P, Jiang Y, Wang X, Wei Q, et al. Disorders of MicroRNAs in peripheral blood mononuclear cells: as novel biomarkers of ankylosing spondylitis and provocative therapeutic targets. Biomed Res Int. 2015;2015:504208. https://doi.org/10.1155/2015/504208.
Wei C, Zhang H, Wei C, Mao Y. Correlation of the expression of miR-146a in peripheral blood mononuclear cells of patients with ankylosing spondylitis and inflammatory factors. Exp Ther Med. 2017;14(5):5027–31. https://doi.org/10.3892/etm.2017.5155.
Ugur M, Baygutalp NK, Melikoglu MA, Baygutalp F, Altas EU, Seferoglu B. Elevated serum interleukin-23 levels in ankylosing spondylitis patients and the relationship with disease activity. Nagoya J Med Sci. 2015;77(4):621–7.
Ranganathan V, Ciccia F, Zeng F, Sari I, Guggino G, Muralitharan J, et al. Macrophage migration inhibitory factor induces inflammation and predicts spinal progression in ankylosing spondylitis. Arthritis Rheumatol. 2017;69(9):1796–806. https://doi.org/10.1002/art.40175.
Qian BP, Ji ML, Qiu Y, Wang B, Yu Y, Shi W, et al. Identification of serum miR-146a and miR-155 as novel noninvasive complementary biomarkers for ankylosing spondylitis. Spine (Phila Pa 1976). 2016;41(9):735–42. https://doi.org/10.1097/BRS.0000000000001339.
Magrey MN, Haqqi T, Haseeb A. Identification of plasma microRNA expression profile in radiographic axial spondyloarthritis-a pilot study. Clin Rheumatol. 2016;35(5):1323–7. https://doi.org/10.1007/s10067-015-3123-7.
• Perez-Sanchez C, Font-Ugalde P, Ruiz-Limon P, Lopez-Pedrera C, Castro-Villegas MC, Abalos-Aguilera MC, et al. Circulating microRNAs as potential biomarkers of disease activity and structural damage in ankylosing spondylitis patients. Hum Mol Genet. 2018;27(5):875–90. https://doi.org/10.1093/hmg/ddy008. This study identified a six-plasma microRNA signature that could be attractive candidates as non-invasive biomarkers for the AS diagnosis.
Cauli A, Shaw J, Giles J, Hatano H, Rysnik O, Payeli S, et al. The arthritis-associated HLA-B*27:05 allele forms more cell surface B27 dimer and free heavy chain ligands for KIR3DL2 than HLA-B*27:09. Rheumatology (Oxford). 2013;52(11):1952–62. https://doi.org/10.1093/rheumatology/ket219.
Zhou L, Zhang Y, Xu H, Hu L, Zhang C, Sun L, et al. Decreased programmed death-1 expression on the T cells of patients with ankylosing spondylitis. Am J Med Sci. 2015;349(6):488–92. https://doi.org/10.1097/MAJ.0000000000000468.
Lee YH, Song GG. Meta-analysis of differentially expressed genes in ankylosing spondylitis. Genet Mol Res. 2015;14(2):5161–70. https://doi.org/10.4238/2015.May.18.6.
•• Chen L, Al-Mossawi MH, Ridley A, Sekine T, Hammitzsch A, de Wit J, et al. miR-10b-5p is a novel Th17 regulator present in Th17 cells from ankylosing spondylitis. Ann Rheum Dis. 2017;76(3):620–5. https://doi.org/10.1136/annrheumdis-2016-210175. First study to determine the miRNA signature in Th-17 cells in AS.
•• Ciccia F, Guggino G, Rizzo A, Saieva L, Peralta S, Giardina A, et al. Type 3 innate lymphoid cells producing IL-17 and IL-22 are expanded in the gut, in the peripheral blood, synovial fluid and bone marrow of patients with ankylosing spondylitis. Ann Rheum Dis. 2015;74(9):1739–47. https://doi.org/10.1136/annrheumdis-2014-206323. This study demonstrated that gut-derived IL-17(+) and IL-22(+)ILC3 expanded in the peripheral blood, SF and inflamed BM of patients with AS suggest the presence of an active homing axis between the gut and the inflamed sacroiliac joints.
Chen D, He J, Lu C, Zhou J, Fang K, Liu X, et al. Increased expression of T cell immunoglobulin and mucin domain 4 is positively associated with the disease severity of patients with ankylosing spondylitis. Inflammation. 2015;38(3):935–40. https://doi.org/10.1007/s10753-014-0055-3.
Akhtari M, Zargar SJ, Mahmoudi M, Vojdanian M, Rezaeimanesh A, Jamshidi A. Ankylosing spondylitis monocyte-derived macrophages express increased level of A2A adenosine receptor and decreased level of ectonucleoside triphosphate diphosphohydrolase-1 (CD39), A1 and A2B adenosine receptors. Clin Rheumatol. 2018;37:1589–95. https://doi.org/10.1007/s10067-018-4055-9.
Smith JA, Barnes MD, Hong D, DeLay ML, Inman RD, Colbert RA. Gene expression analysis of macrophages derived from ankylosing spondylitis patients reveals interferon-gamma dysregulation. Arthritis Rheum. 2008;58(6):1640–9. https://doi.org/10.1002/art.23512.
Zhu ZQ, Tang JS, Cao XJ. Transcriptome network analysis reveals potential candidate genes for ankylosing spondylitis. Eur Rev Med Pharmacol Sci. 2013;17(23):3178–85.
Bhattacharya A, Eissa NT. Autophagy and autoimmunity crosstalks. Front Immunol. 2013;4:88. https://doi.org/10.3389/fimmu.2013.00088.
Cai A, Qi S, Su Z, Shen H, Yang Y, He L, et al. Quantitative proteomic analysis of peripheral blood mononuclear cells in ankylosing spondylitis by iTRAQ. Clin Transl Sci. 2015;8(5):579–83. https://doi.org/10.1111/cts.12265.
Neerinckx B, Carter S, Lories R. IL-23 expression and activation of autophagy in synovium and PBMCs of HLA-B27 positive patients with ankylosing spondylitis. Response to: ‘evidence that autophagy, but not the unfolded protein response, regulates the expression of IL-23 in the gut of patients with ankylosing spondylitis and subclinical gut inflammation’ by Ciccia et al. Ann Rheum Dis. 2014;73(11):e68. https://doi.org/10.1136/annrheumdis-2014-206277.
Wang Y, Luo J, Wang X, Yang B, Cui L. MicroRNA-199a-5p induced autophagy and inhibits the pathogenesis of ankylosing spondylitis by modulating the mTOR signaling via directly targeting Ras homolog enriched in brain (Rheb). Cell Physiol Biochem. 2017;42(6):2481–91. https://doi.org/10.1159/000480211.
Xia Y, Chen K, Zhang MH, Wang LC, Ma CY, Lin YL, et al. MicroRNA-124 involves in ankylosing spondylitis by targeting ANTXR2. Mod Rheumatol. 2015;25(5):784–9. https://doi.org/10.3109/14397595.2015.1023887.
Neerinckx B, Lories R. Mechanisms, impact and prevention of pathological bone regeneration in spondyloarthritis. Curr Opin Rheumatol. 2017;29(4):287–92. https://doi.org/10.1097/BOR.0000000000000404.
Shaw AT, Gravallese EM. Mediators of inflammation and bone remodeling in rheumatic disease. Semin Cell Dev Biol. 2016;49:2–10. https://doi.org/10.1016/j.semcdb.2015.10.013.
Yang Y, Dai M. Expression of PADI4 in patients with ankylosing spondylitis and its role in mediating the effects of TNF-alpha on the proliferation and osteogenic differentiation of human mesenchymal stem cells. Int J Mol Med. 2015;36(2):565–70. https://doi.org/10.3892/ijmm.2015.2248.
Peng W, Zhu S, Li X, Weng J, Chen S. miR-27b-3p suppressed osteogenic differentiation of maxillary sinus membrane stem cells by targeting Sp7. Implant Dent. 2017;26(4):492–9. https://doi.org/10.1097/ID.0000000000000637.
Fang T, Wu Q, Zhou L, Mu S, Fu Q. miR-106b-5p and miR-17-5p suppress osteogenic differentiation by targeting Smad5 and inhibit bone formation. Exp Cell Res. 2016;347(1):74–82. https://doi.org/10.1016/j.yexcr.2016.07.010.
Chen MH, Chen HA, Chen WS, Chen MH, Tsai CY, Chou CT. Upregulation of BMP-2 expression in peripheral blood mononuclear cells by proinflammatory cytokines and radiographic progression in ankylosing spondylitis. Mod Rheumatol. 2015;25(6):913–8. https://doi.org/10.3109/14397595.2015.1029221.
Yuan B, Wu Z. MMP-2 silencing reduces the osteogenic transformation of fibroblasts by inhibiting the activation of the BMP/Smad pathway in ankylosing spondylitis. Oncol Lett. 2018;15(3):3281–6. https://doi.org/10.3892/ol.2017.7714.
Liu KG, He QH, Tan JW, Liao GJ. Expression of TNF-alpha, VEGF, and MMP-3 mRNAs in synovial tissues and their roles in fibroblast-mediated osteogenesis in ankylosing spondylitis. Genet Mol Res. 2015;14(2):6852–8. https://doi.org/10.4238/2015.June.18.28.
Poddubnyy D, Conrad K, Haibel H, Syrbe U, Appel H, Braun J, et al. Elevated serum level of the vascular endothelial growth factor predicts radiographic spinal progression in patients with axial spondyloarthritis. Ann Rheum Dis. 2014;73(12):2137–43. https://doi.org/10.1136/annrheumdis-2013-203824.
Daoussis D, Filippopoulou A, Liossis SN, Sirinian C, Klavdianou K, Bouris P, et al. Anti-TNFalpha treatment decreases the previously increased serum Indian hedgehog levels in patients with ankylosing spondylitis and affects the expression of functional hedgehog pathway target genes. Semin Arthritis Rheum. 2015;44(6):646–51. https://doi.org/10.1016/j.semarthrit.2015.01.004.
Heiland GR, Appel H, Poddubnyy D, Zwerina J, Hueber A, Haibel H, et al. High level of functional dickkopf-1 predicts protection from syndesmophyte formation in patients with ankylosing spondylitis. Ann Rheum Dis. 2012;71(4):572–4. https://doi.org/10.1136/annrheumdis-2011-200216.
Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem. 2009;108(1):216–24. https://doi.org/10.1002/jcb.22243.
Li DH, Fu HD, Liu WL, Wei YS, Jiang DM. Decreased local and systematic matrix Gla protein (MGP) expression and its link to radiographic progression in ankylosing spondylitis patients. Clin Lab. 2015;61(3–4):243–9.
Mann M, Barad O, Agami R, Geiger B, Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc Natl Acad Sci U S A. 2010;107(36):15804–9. https://doi.org/10.1073/pnas.0915022107.
• Prajzlerova K, Grobelna K, Husakova M, Forejtova S, Jungel A, Gay S, et al. Association between circulating miRNAs and spinal involvement in patients with axial spondyloarthritis. PLoS One. 2017;12(9):e0185323. https://doi.org/10.1371/journal.pone.0185323. This study demonstrated that circulating miRNAs play a role in the pathogenesis and progression of AxSpA.
Taylan A, Sari I, Akinci B, Bilge S, Kozaci D, Akar S, et al. Biomarkers and cytokines of bone turnover: extensive evaluation in a cohort of patients with ankylosing spondylitis. BMC Musculoskelet Disord. 2012;13:191. https://doi.org/10.1186/1471-2474-13-191.
Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117(13):3648–57. https://doi.org/10.1182/blood-2010-10-311415.
Huang CH, Wei JC, Chang WC, Chiou SY, Chou CH, Lin YJ, et al. Higher expression of whole blood microRNA-21 in patients with ankylosing spondylitis associated with programmed cell death 4 mRNA expression and collagen cross-linked C-telopeptide concentration. J Rheumatol. 2014;41(6):1104–11. https://doi.org/10.3899/jrheum.130515.
• Li C, Zhang P, Gu J. miR-29a modulates tumor necrosis factor-alpha-induced osteogenic inhibition by targeting Wnt antagonists. Develop Growth Differ. 2015;57(3):264–73. https://doi.org/10.1111/dgd.12207. This study determiend that a specific miRNA regulates tumor necrosis factor-α (TNF-α) mediated bone loss through activating the Wnt/β-catenin pathway.
Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature. 2012;491(7424):384–92. https://doi.org/10.1038/nature11708.
• Menni C, Zierer J, Valdes AM, Spector TD. Mixing omics: combining genetics and metabolomics to study rheumatic diseases. Nat Rev Rheumatol. 2017;13(3):174–81. https://doi.org/10.1038/nrrheum.2017.5. Insightful review of the potential for combining genetics and metabolomics to elucidate novel markers for precision medicine.
Zhai G, Randell EW, Rahman P. Metabolomics of osteoarthritis: emerging novel markers and their potential clinical utility. Rheumatology (Oxford). 2018; https://doi.org/10.1093/rheumatology/kex497.
•• Wang W, Yang GJ, Zhang J, Chen C, Jia ZY, Li J, et al. Plasma, urine and ligament tissue metabolite profiling reveals potential biomarkers of ankylosing spondylitis using NMR-based metabolic profiles. Arthritis Res Ther. 2016;18(1):244. https://doi.org/10.1186/s13075-016-1139-2. 1H NMR-based metabolic profiling on plasma, urine and ligament implicating fat metabolism and intestinal microbial metabolism.
• Gao P, Lu C, Zhang F, Sang P, Yang D, Li X, et al. Integrated GC-MS and LC-MS plasma metabonomics analysis of ankylosing spondylitis. Analyst. 2008;133(9):1214–20. https://doi.org/10.1039/b807369d. MS-based metabolomics approach that determined plasma glucose, phosphate, phenylanalyine and phospholipids were significantly altered in AS patients.
•• Stoll ML, Kumar R, Lefkowitz EJ, Cron RQ, Morrow CD, Barnes S. Fecal metabolomics in pediatric spondyloarthritis implicate decreased metabolic diversity and altered tryptophan metabolism as pathogenic factors. Genes Immun. 2016;17(7):400–5. https://doi.org/10.1038/gene.2016.38. Global metabolic profiling of fecal samples revealed decreased metabolic diversity and altered tryptophan metabolism in early and late JIA/ERA cohorts.
Uddin M, Codner D, Hasan SM, Scherer SW, O'Rielly DD, Rahman P. Integrated genomics identifies convergence of ankylosing spondylitis with global immune mediated disease pathways. Sci Rep. 2015;5:10314. https://doi.org/10.1038/srep10314.
Yang Y, Wang L, Wang S, Huang R, Zheng L, Liang S, et al. An integrated metabonomic approach to studying metabolic profiles in rat models with insulin resistance induced by high fructose. Mol BioSyst. 2014;10(7):1803–11. https://doi.org/10.1039/c3mb70618d.
Pallister T, Jackson MA, Martin TC, Zierer J, Jennings A, Mohney RP, et al. Hippurate as a metabolomic marker of gut microbiome diversity: modulation by diet and relationship to metabolic syndrome. Sci Rep. 2017;7(1):13670. https://doi.org/10.1038/s41598-017-13722-4.
Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517–26. https://doi.org/10.1016/j.cmet.2011.02.018.
Roediger WE, Nance S. Selective reduction of fatty acid oxidation in colonocytes: correlation with ulcerative colitis. Lipids. 1990;25(10):646–52.
O'Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res. 2015;277:32–48. https://doi.org/10.1016/j.bbr.2014.07.027.
Etienne-Mesmin L, Chassaing B, Gewirtz AT. Tryptophan: a gut microbiota-derived metabolites regulating inflammation. World J Gastrointest Pharmacol Ther. 2017;8(1):7–9. https://doi.org/10.4292/wjgpt.v8.i1.7.
Kaur H, Edmonds SE, Blake DR, Halliwell B. Hydroxyl radical generation by rheumatoid blood and knee joint synovial fluid. Ann Rheum Dis. 1996;55(12):915–20.
Haugen MA, Kjeldsen-Kragh J, Bjerve KS, Hostmark AT, Forre O. Changes in plasma phospholipid fatty acids and their relationship to disease activity in rheumatoid arthritis patients treated with a vegetarian diet. Br J Nutr. 1994;72(4):555–66.
Buchman AL, Dubin MD, Moukarzel AA, Jenden DJ, Roch M, Rice KM, et al. Choline deficiency: a cause of hepatic steatosis during parenteral nutrition that can be reversed with intravenous choline supplementation. Hepatology. 1995;22(5):1399–403.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Dr. Rahman reports personal fees from Abbott, AbbVie, Amgen, Celgene, Eli Lilly, Novartis, Pfizer, Roche, and UCB grants and personal fees from Janssen, outside the submitted work.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Competing Interests
PR is a consultant to multiple pharmaceutical companies dealing with biologic agents including Abbott, AbbVie, Amgen, Celgene, Eli Lilly, Janssen, Novartis, Pfizer, Roche, and UCB,
Additional information
This article is part of the Topical Collection on Spondyloarthritis
Rights and permissions
About this article
Cite this article
O’Rielly, D.D., Zhai, G. & Rahman, P. Expression and Metabolomic Profiling in Axial Spondyloarthritis. Curr Rheumatol Rep 20, 51 (2018). https://doi.org/10.1007/s11926-018-0756-y
Published:
DOI: https://doi.org/10.1007/s11926-018-0756-y