Abstract
Recent advances in massively parallel sequencing (MPS) have had an extensive impact on research in medical genomics. In particular, the analysis of rare variants using MPS promises to lead to a better understanding of complex disorders. Nevertheless, for meaningful studies that address the genetic basis for neuropsychiatric disorders, at least hundreds of patient samples have to be analyzed. This undertaking is still not feasible for single research groups on a whole-genome scale and in individual samples. Thus, researchers increasingly employ strategies for reducing the amount of sequencing efforts, such as target enrichment and non-barcoded sample pooling. This review provides an overview of current technologies, discusses options for reduced experimental designs, and illustrates the successful application of the presented methodologies in a recent study of panic disorder patients. Thereby, it aims to introduce the emerging field of MPS into neuropsychiatric research and might serve as a guide for further studies.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
A Catalog of Published Genome-Wide Association Studies [database on the Internet]. Available from: http://www.genome.gov/gwastudies/. Accessed: August 8th 2012.
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi:10.1038/nature08494.
Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet. 2008;9(4):255–66. doi:10.1038/nrg2322.
Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 2001;158(10):1568–78.
Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–52. doi:10.1038/nature08185.
Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456(7218):18–21. doi:10.1038/456018a. Review that points out limitations of published GWAS in common diseases. It introduces to the phenomenon of missing heritability and presents explanation models.
Frank RA, McRae AF, Pocklington AJ, van de Lagemaat LN, Navarro P, Croning MD, et al. Clustered coding variants in the glutamate receptor complexes of individuals with schizophrenia and bipolar disorder. PLoS One. 2011;6(4):e19011. doi:10.1371/journal.pone.0019011PONE-D-10-05384.
Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42(8):684–7. doi:10.1038/ng.628.
Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43(12):1193–201. doi:10.1038/ng.998.
Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–45. doi:10.1038/nbt1486.
Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–45. doi:10.1038/nature03001.
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi:10.1038/35057062.
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi:10.1126/science.1058040291/5507/1304.
Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11(1):31–46. doi:10.1038/nrg2626. A detailed overview over the most used massively parallel sequencing platforms, working principles, and their applications.
Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11(5):473–83. doi:10.1093/bib/bbq015. The authors review currently available mapping methods for MPS data.
Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329(5987):75–8. doi:10.1126/science.1190371.
Stratton M. Genome resequencing and genetic variation. Nat Biotechnol. 2008;26(1):65–6. doi:10.1038/nbt0108-65.
Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, et al. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7(2):111–8. doi:10.1038/nmeth.1419. This review provides a quick and profound overview over target enrichment strategies and some ideas about choosing the most suitable technology.
Chen X, Listman JB, Slack FJ, Gelernter J, Zhao H. Biases and errors on allele frequency estimation and disease association tests of next-generation sequencing of pooled samples. Genet Epidemiol. 2012. doi:10.1002/gepi.21648.
Altmann A, Weber P, Bader D, Preuß M, Binder EB, Müller-Myhsok B. A beginners guide To SNP calling from high-throughput DNA-sequencing data. Hum Gen. 2012;131(10):1541–54. doi:10.1007/s00439-012-1213-z.
Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12(6):443–51. doi:10.1038/nrg2986. A review on currently available SNP calling methods in non-pooled DNA.
Vallania FL, Druley TE, Ramos E, Wang J, Borecki I, Province M, et al. High-throughput discovery of rare insertions and deletions in large cohorts. Genome Res. 2010;20(12):1711–8. doi:10.1101/gr.109157.110.
Bansal V. A statistical method for the detection of variants from next-generation resequencing of DNA pools. Bioinformatics. 2010;26(12):i318–24. doi:10.1093/bioinformatics/btq214.
Altmann A, Weber P, Quast C, Rex-Haffner M, Binder EB, Muller-Myhsok B. vipR: variant identification in pooled DNA using R. Bioinformatics. 2011;27(13):i77–84. doi:10.1093/bioinformatics/btr205.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi:10.1093/nar/gkq603.
Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34(Web Server issue):W635–41. doi:10.1093/nar/gkl236.
Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–41. doi:10.1101/gr.77240313/9/2129.
Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32(8):894–9.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. doi:10.1038/nprot.2009.86.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi:10.1101/gr.229102.
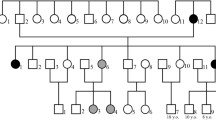
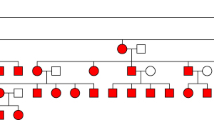
Quast C, Altmann A, Weber P, Arloth J, Bader D, Heck A et al. Rare variants in TMEM132D in a case-control sample for panic disorder. Am J Med Genet B Neuropsychiatr Genet. 2012;(in press).
Erhardt A, Czibere L, Roeske D, Lucae S, Unschuld PG, Ripke S, et al. TMEM132D, a new candidate for anxiety phenotypes: evidence from human and mouse studies. Mol Psychiatr. 2010;16(6):647–63. doi:10.1038/mp.2010.41.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi:10.1093/bioinformatics/btp324.
Bansal V, Libiger O, Torkamani A, Schork NJ. Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet. 2010;11(11):773–85. doi:10.1038/nrg2867. An excellent overview about the statistical challenges when working with rare genetic variants.
Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST). Mutat Res. 2007;615(1–2):28–56. doi:10.1016/j.mrfmmm.2006.09.003.
Disclosure
No potential conflicts of interest relevant to this article were reported.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Genetic Disorders
Rights and permissions
About this article
Cite this article
Altmann, A., Quast, C. & Weber, P. Detecting Rare Variants for Psychiatric Disorders Using Next Generation Sequencing: A Methods Primer. Curr Psychiatry Rep 15, 333 (2013). https://doi.org/10.1007/s11920-012-0333-4
Published:
DOI: https://doi.org/10.1007/s11920-012-0333-4
Keywords
- Next-generation sequencing
- NGS
- Massively parallel sequencing
- MPS
- Targeted re-sequencing
- Target enrichment
- Exome enrichment
- Sample pooling
- DNA pooling
- Rare genetic variants
- Bioinformatics
- Complex disease
- Psychiatric disorder
- Anxiety disorder
- Panic disorder
- Human genetics
- Genetic disorders
- Genome-wide association studies
- GWAS
- Example study
- Review
- Psychiatry