Abstract
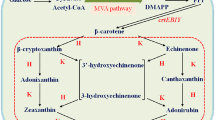
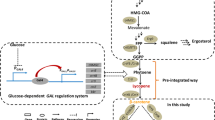
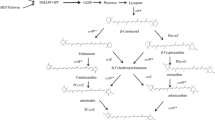
The conversion of β-carotene to astaxanthin is a complex pathway network, in which two steps of hydroxylation and two steps of ketolation are catalyzed by β-carotene hydroxylase (CrtZ) and β-carotene ketolase (CrtW) respectively. Here, astaxanthin biosynthesis pathway was constructed in Saccharomyces cerevisiae by introducing heterologous CrtZ and CrtW into an existing high β-carotene producing strain. Both genes crtZ and crtW were codon optimized and expressed under the control of constitutive promoters. Through combinatorial expression of CrtZ and CrtW from diverse species, nine strains in dark red were visually chosen from thirty combinations. In all the selected strains, strain SyBE_Sc118060 with CrtW from Brevundimonas vesicularis DC263 and CrtZ from Alcaligenes sp. strain PC-1 achieved the highest astaxanthin yield of 3.1 mg/g DCW. Protein phylogenetic analysis shows that the shorter evolutionary distance of CrtW is, the higher astaxanthin titer is. Further, when the promoter of crtZ in strain SyBE_Sc118060 was replaced from FBA1p to TEF1p, the astaxanthin yield was increased by 30.4% (from 3.4 to 4.5 mg/g DCW). In the meanwhile, 33.5-fold increase on crtZ transcription level and 39.1-fold enhancement on the transcriptional ratio of crtZ to crtW were observed at early exponential phase in medium with 4% (w/v) glucose. Otherwise, although the ratio of crtZ to crtW were increased at mid-, late-exponential phases in medium with 2% (w/v) glucose, the transcription level of both crtZ and crtW were actually decreased during the whole time course, consequently leading to no significant improvement on astaxanthin production. Finally, through high cell density fed-batch fermentation using a carbon source restriction strategy, the production of astaxanthin in a 5-L bioreactor reached to 81.0 mg/L, which was the highest astaxanthin titer reported in yeast. This study provides a reference to greatly enhance desired compounds accumulation by employing the key enzyme(s) in microbes.

Similar content being viewed by others
References
Ambati R R, Phang S M, Ravi S, Aswathanarayana R G. Astaxanthin: sources, extraction, stability, biological activities and its commercial applications—a review. Marine Drugs, 2014, 12(1): 128–152
Zhou P, Ye L, Xie W, Lv X, Yu H. Highly efficient biosynthesis of astaxanthin in Saccharomyces cerevisiae by integration and tuning of algal crtZ and bkt. Applied Microbiology and Biotechnology, 2015, 99(20): 8419–8428
Ukibe K, Hashida K, Yoshida N, Takagi H. Metabolic engineering of Saccharomyces cerevisiae for astaxanthin production and oxidative stress tolerance. Applied and Environmental Microbiology, 2009, 75(22): 7205–7211
Martin J F, Gudina E, Barredo J L. Conversion of beta-carotene into astaxanthin: Two separate enzymes or a bifunctional hydroxylaseketolase protein. Microbial Cell Factories, 2008, 7(1): 3
Chang J J, Thia C, Lin H Y, Liu H L, Ho F J, Wu J T, Shih M C, Li W H, Huang C C. Integrating an algal beta-carotene hydroxylase gene into a designed carotenoid-biosynthesis pathway increases carotenoid production in yeast. Bioresource Technology, 2015, 184: 2–8
Sarria S, Wong B, Garcia M H, Keasling J D, Peralta-Yahya P. Microbial synthesis of pinene. ACS Synthetic Biology, 2014, 3(7): 466–475
Chen Y, Xiao W, Wang Y, Liu H, Li X, Yuan Y. Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering. Microbial Cell Factories, 2016, 15(1): 113
Du H X, Xiao W H, Wang Y, Zhou X, Zhang Y, Liu D, Yuan Y J. Engineering Yarrowia lipolytica for campesterol overproduction. PLoS One, 2016, 11(1): e0146773
Fraser P D, Miura Y, Misawa N. In vitro characterization of astaxanthin biosynthetic enzymes. Journal of Biological Chemistry, 1997, 272(10): 6128–6135
Ye R W, Stead K J, Yao H, He H. Mutational and functional analysis of the beta-carotene ketolase involved in the production of canthaxanthin and astaxanthin. Applied and Environmental Microbiology, 2006, 72(9): 5829–5837
Tao L, Wilczek J, Odom J M, Cheng Q. Engineering a beta-carotene ketolase for astaxanthin production. Metabolic Engineering, 2006, 8(6): 523–531
Scaife M A, Burja A M, Wright P C. Characterization of cyanobacterial beta-carotene ketolase and hydroxylase genes in Escherichia coli, and their application for astaxanthin biosynthesis. Biotechnology and Bioengineering, 2009, 103(5): 944–955
Choi S K, Nishida Y, Matsuda S, Adachi K, Kasai H, Peng X, Komemushi S, Miki W, Misawa N. Characterization of betacarotene ketolases, CrtW, from marine bacteria by complementation analysis in Escherichia coli. Marine Biotechnology (New York, N. Y.), 2005, 7(5): 515–522
Fraser P D, Shimada H, Misawa N. Enzymic confirmation of reactions involved in routes to astaxanthin formation, elucidated using a direct substrate in vitro assay. European Journal of Biochemistry, 1998, 252(2): 229–236
Zelcbuch L, Antonovsky N, Bar-Even A, Levin-Karp A, Barenholz U, Dayagi M, Liebermeister W, Flamholz A, Noor E, Amram S, et al. Spanning high-dimensional expression space using ribosomebinding site combinatorics. Nucleic Acids Research, 2013, 41(9): e98
Ajikumar P K, Xiao W H, Tyo K E, Wang Y, Simeon F, Leonard E, Mucha O, Phon T H, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science, 2010, 330(6000): 70–74
Cao Y X, Xiao W H, Liu D, Zhang J L, Ding M Z, Yuan Y J. Biosynthesis of odd-chain fatty alcohols in Escherichia coli. Metabolic Engineering, 2015, 29: 113–123
Lemuth K, Steuer K, Albermann C. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microbial Cell Factories, 2011, 10(1): 29
Wang R Z, Pan C H, Wang Y, Xiao W H, Yuan Y J. Design and construction of high β-carotene producing Saccharomyces cerevisiae. Journal of Chinese Biotechnology, 2016, 36: 83–91
Gietz R D, Schiestl R H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols, 2007, 2(1): 31–34
Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 2016, 33(7): 1870–1874
Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 1987, 4: 406–425
Sanderson M J, Wojciechowski M F. Improved bootstrap confidence limits in large-scale phylogenies, with an example from neo-astragalus (Leguminosae). Systematic Biology, 2000, 49: 671–685
Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel H J, eds. Evolving Genes and Proteins. New York: Academic Press, 1965, 97–166
Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (San Diego, Calif.), 2001, 25(4): 402–408
Jin Z, Wong A, Foo J L, Ng J, Cao Y X, Chang M W, Yuan Y J. Engineering Saccharomyces cerevisiae to produce odd chain-length fatty alcohols. Biotechnology and Bioengineering, 2016, 113(4): 842–851
Lorenz R T, Cysewski G R. Commercial potential for Haematococcus microalgae as a natural source of astaxanthin. Trends in Biotechnology, 2000, 18(4): 160–167
Scaife M A, Ma C A, Ninlayarn T, Wright P C, Armenta R E. Comparative analysis of beta-carotene hydroxylase genes for astaxanthin biosynthesis. Journal of Natural Products, 2012, 75(6): 1117–1124
Nam H, Lewis N E, Lerman J A, Lee D H, Chang R L, Kim D, Palsson B O. Network context and selection in the evolution to enzyme specificity. Science, 2012, 337(6098): 1101–1104
Sandmann G. Evolution of carotene desaturation: The complication of a simple pathway. Archives of Biochemistry and Biophysics, 2009, 483(2): 169–174
Schaub P, Yu Q, Gemmecker S, Poussin-Courmontagne P, Mailliot J, McEwen A G, Ghisla S, Al-Babili S, Cavarelli J, Beyer P. On the structure and function of the phytoene desaturase CrtI from Pantoea ananatis, a membrane-peripheral and FAD-dependent oxidase/ isomerase. PLoS One, 2012, 7(6): e39550
Sun J, Shao Z, Zhao H, Nair N, Wen F, Xu J H, Zhao H. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnology and Bioengineering, 2012, 109(8): 2082–2092
Peng B, Williams T C, Henry M, Nielsen L K, Vickers C E. Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: A comparison of yeast promoter activities. Microbial Cell Factories, 2015, 14(1): 91
Acknowledgements
This work was supported by the International S&T Cooperation Program of China (2015DFA00960), the National Natural Science Foundation of China (Grant Nos. 31600052 and 21676192) and Innovative Talents and Platform Program of Tianjin (16PTSYJC00050).
Author information
Authors and Affiliations
Corresponding authors
Additional information
These authors contributed equally to this work
Electronic Supplementary Material Supplementary material is available in the online version of this article at http://dx.doi.org/10.1007/s11705-017-1628-0 and is accessible for authorized users.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wang, R., Gu, X., Yao, M. et al. Engineering of β-carotene hydroxylase and ketolase for astaxanthin overproduction in Saccharomyces cerevisiae. Front. Chem. Sci. Eng. 11, 89–99 (2017). https://doi.org/10.1007/s11705-017-1628-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11705-017-1628-0