Abstract
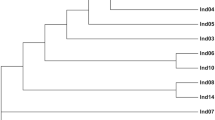
Iranian Bactrian camel population is less than 100 animals. Iranian biological resource center produced more than 50 Bactrian camel fibroblast cell lines as a somatic cell bank for conservation animal genetic resources. We compared two type markers performance, including 14 random amplified polymorphic DNA (RAPDs) (dominant) and eight microsatellite (co-dominant) for cell line identification, individual identification and investigation genetic structure of these samples. Based on clarity, polymorphism, and repeatability, four RAPD primers were selected for future analysis. Four RAPD primers and eight microsatellite markers have generated a total of 21 fragments and 45 alleles, respectively. RAPD primers revealed fragment size between 150 to 2000 bp and gene diversity since 0.27 (IBRD) to 0.46 (GC10), with an average of 0.37. Microsatellite markers generated number of alleles per locus ranged from 3 to 11, with an average of 5.62 alleles. The observed heterozygosity ranged from 0.359 (IBRC02) to 0.978 (YWLL08), and expected heterozygosity ranged from 0.449 (IBRC02) to 0.879 (YWLL08). Bottleneck analysis and curve showed that Bactrian camel population did not experience a low diversity. RAPD profiles were especially suitable for investigation population genetics. All primers generated novel and polymorphic fragments. Briefly, our results show that a multiplex PCR based on these markers can still be valuable and suitable for authentication of cell lines, investigating gene diversity and conservation genetic resources in Bactrian camel, while new technologies are continuously developed.


Similar content being viewed by others
References
Al-Atiyat RM (2015) The power of 28 microsatellite markers for parentage testing in sheep. Electron J Biotechnol 18(2):116–121. https://doi.org/10.1016/j.ejbt.2015.01.001
Almeida JL, Hill CR, Cole KD (2011) Authentication of African green monkey cell lines using human short tandem repeat markers. BMC Biotechnol 11(1):102. https://doi.org/10.1186/1472-6750-11-102
Amoli AD, Aminafshar M, Fazeli SS, Emam N, Kashan J, Khaledi KJ (2017a) Isolation and characterization of microsatellite markers from endangered species (Camelus bactrianus). Iran J Appl Anim Sci 7(4):693–698
Amoli AD, Mohebali N, Farzaneh P, Fazeli SAS, Nikfarjam L, Movasagh SA, Moradmand Z, Ganjibakhsh M, Nasimian A, Izadpanah M (2017b) Establishment and characterization of Caspian horse fibroblast cell bank in Iran. In Vitro Cell Dev Biol Anim 53(4):337–343. https://doi.org/10.1007/s11626-016-0120-3
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144(4):2001–2014
Cornuet J-M, Piry S, Luikart G, Estoup A, Solignac M (1999) New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153(4):1989–2000
Cristescu R, Sherwin WB, Handasyde K, Cahill V, Cooper DW (2010) Detecting bottlenecks using BOTTLENECK 1.2.02 in wild populations: the importance of the microsatellite structure. Conserv Genet 11(3):1043–1049
Dittmann MT, Runge U, Lang RA, Moser D, Galeffi C, Kreuzer M, Clauss M (2014) Methane emission by camelids. PLoS One 9(4):e94363
Easley CA 4th, Phillips BT, McGuire MM, Barringer JM, Valli H, Hermann BP, Simerly CR, Rajkovic A, Miki T, Orwig KE, GP S (2012) Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep 2(3):440–446. https://doi.org/10.1016/j.celrep.2012.07.015
Finger A, Klank C (2010). Review molecular methods: blessing or curse? Relict Species, Springer: 309–320, DOI: https://doi.org/10.1007/978-3-540-92160-8_18
Freshney RI (2005) Culture of specific cell types. Wiley Online Library
Gorji ZE, Khaledi KJ, Amoli AD, Ganjibakhsh M, Nasimian A, Gohari NS, Izadpanah M, Vakhshiteh F, Farghadan M, Moghanjoghi SM (2016) Establishment and characteristics of Iranian Sistani cattle fibroblast bank: a way to genetic conservation. Conserv Genet Resour 9(2):305–312
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16(5):1099–1106
Kuz’mina E E (2008) The prehistory of the silk road. University of Pennsylvania Press
Li LF, Guan WJ, Li H, Zhou XZ, Bai XJ, Ma YH (2009) Establishment and characterization of a fibroblast cell line derived from Texel sheep. Biochem Cell Biol 87(3):485–492. https://doi.org/10.1139/O09-005
Mahrous KF, Ramadan HA, Abdel-Aziem SH, Abd-El Mordy M, Hemdan DM (2011) Genetic variations between camel breeds using microsatellite markers and RAPD techniques. J. Appl. Biosci 39:2626–2634
Makkar HP, Viljoen GJ (2005) Applications of gene-based technologies for improving animal production and health in developing countries. Springer. doi: https://doi.org/10.1007/b105256
Mehta S, Mishra B, Sahani M (2006) Genetic differentiation of Indian camel (Camelus dromedarius) breeds using random oligonucleotide primers. Animal Genetic Resources/Resources génétiques animales/Recursos genéticos animales 39(77–88
Nagy S, Poczai P, Cernák I, Gorji AM, Hegedűs G, Taller J (2012) PICcalc: an online program to calculate polymorphic information content for molecular genetic studies. Biochem Genet 50(9–10):670–672. https://doi.org/10.1007/s10528-012-9509-1
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89(3):583–590
Pavel AB, Vasile CI (2012) PyElph-a software tool for gel images analysis and phylogenetics. BMC Bioinformatics 13(1):9. https://doi.org/10.1186/1471-2105-13-9
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Roosen J, Fadlaoui A, Bertaglia M (2005) Economic evaluation for conservation of farm animal genetic resources. J Anim Breed Genet 122(4):217–228
Shahkarami S, Afraz F, Sayed MZ, Banabazi M, Asadzadeh N, Asadi N, Hemmati B, Ghanbari A, Razavi K (2012) Genetic diversity in Iranian Bactrian camels (Camelus batrianus) using, microsatellite markers. Mod Genet J 7(330):249–258
Stacey G (2000) Cell contamination leads to inaccurate data: we must take action now. Nature 403(6768):356–356. https://doi.org/10.1038/35000394
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676. https://doi.org/10.1016/j.cell.2006.07.024
Turlure C, Vandewoestijne S, Baguette M (2014) Conservation genetics of a threatened butterfly: comparison of allozymes, RAPDs and microsatellites. BMC Genet 15(1):114
Vandewoestijne S, Schtickzelle N, Baguette M (2008) Positive correlation between genetic diversity and fitness in a large, well-connected metapopulation. BMC Biol 6(1):46
Wu H, Guang X, Al-Fageeh MB, Cao J, Pan S, Zhou H, Zhang L, Abutarboush MH, Xing Y, Xie Z (2014) Camelid genomes reveal evolution and adaptation to desert environments. Nat Commun 5:5188. https://doi.org/10.1038/ncomms6188
Yeh FC, Yang R-C, Boyle T (1999) POPGENE version 1.31. Microsoft window-based freeware for population genetic analysis. University of Alberta, Edmonton
Acknowledgements
The authors express their gratitude to Parvaneh Farzaneh for providing technical advice and access to required equipment and all colleagues in Human and Animal cell bank.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Editor: Tetsuji Okamoto
Rights and permissions
About this article
Cite this article
Daneshvar Amoli, A., Shahzadeh Fazeli, S.A., Aminafshar, M. et al. Identification of Bactrian camel cell lines using genetic markers. In Vitro Cell.Dev.Biol.-Animal 54, 265–271 (2018). https://doi.org/10.1007/s11626-018-0238-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-018-0238-6