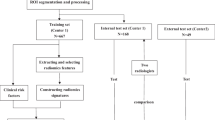
Abstract
Purpose
To compare texture feature estimates obtained from 18F-FDG-PET images using three different software packages.
Methods
PET images from 15 patients with head and neck cancer were processed with three different freeware software: CGITA, LIFEx, and Metavol. For each lesion, 38 texture features were extracted from each software package. To evaluate the statistical agreement among the features across packages a non-parametric Kruskal–Wallis test was used. Differences in the features between each couple of software were assessed using a subsequent Dunn test. Correlation between texture features was evaluated via the Spearman coefficient.
Results
Twenty-three of 38 features showed a significant agreement across the three software (P < 0.05). The agreement was better between LIFEx vs. Metavol (36 of 38) and worse between CGITA and Metavol (24 of 38), and CGITA vs. LIFEx (23 of 38). All features resulted correlated (ρ > = 0.70, P < 0.001) in comparing LIFEx vs. Metavol. Seven of 38 features were found not in agreement and slightly or not correlated (ρ < 0.70, P < 0.001) in comparing CGITA vs. LIFEx, and CGITA vs. Metavol.
Conclusion
Some texture discrepancies across software packages exist. Our findings reinforce the need to continue the standardization process, and to succeed in building a reference dataset to be used for comparisons.





Similar content being viewed by others
References
Gonzalez RC, Woods RE. Digital image processing. Truskey: Third Edition - Pearson Prentice Hall; 2008. p. 827–56.
Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;3(5):4006.
Heppner GH. Tumor heterogeneity. Cancer Res. 1984;44(6):2259–65.
Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15(2):81–94.
Kitajima K, Suenaga Y, Sugimura K. Present and future role of FDG-PET/CT imaging in the management of head and neck carcinoma. Jpn J Radiol. 2015;33(12):776–89.
Buvat I, Orlhac F, Soussan M. Tumor texture analysis in pet: where do we stand. J Nucl Med. 2015;56(11):1642–4.
O’Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21(2):249–57.
Nyflot MJ, Yang F, Byrd D, Bowen SR, Sandison GA, Kinahan PE. Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. J Med Imaging (Bellingham). 2015;2(4):041002.
Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D. Characterization of PET/CT images using texture analysis: the past, the present… any future. Eur J Nucl Med Mol Imaging. 2017;44(1):151–65.
van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11(1):91.
Image biomarker standardization initiative reference manual. https://arxiv.org/pdf/1612.07003.pdf. Accessed 2 Oct 2020.
Zwanenburg A. Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging. 2019;46(13):2638–55.
Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–38.
Fang YH, Lin CY, Shih MJ, Wang HM, Ho TY, Liao CT, et al. Development and evaluation of an open-source software package “CGITA” for quantifying tumor heterogeneity with molecular images. Biomed Res Int. 2014;2014:248505.
Nioche C, Orlhac F, Boughdad S, Reuzé S, Goya-Outi J, Robert C, et al. LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018;78(16):4786–9.
Hirata K, Kobayashi K, Wong KP, Manabe O, Surmak A, Tamaki N, et al. A semi-automated technique determining the liver standardized uptake value reference for tumor delineation in FDG PET-CT. PLoS ONE. 2014;9(8):e105682.
van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–7.
Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys. 2015;42(3):1341–53.
Apte AP, Iyer A, Crispin-Ortuzar M, Pandya R, van Dijk LV, Spezi E, et al. Technical note: extension of CERR for computational radiomics: a comprehensive MATLAB platform for reproducible radiomics research. Med Phys. 2015. https://doi.org/10.1002/mp.13046.
Foy JJ, Robinson KR, Li H, Giger ML, Al-Hallaq H, Armato SG. Variation in algorithm implementation across radiomics software. J Med Imaging (Bellingham). 2018;5(4):044505.
Liang ZG, Tan HQ, Zhang F, Rui Tan LK, Lin L, Lenkowicz J, et al. Comparison of radiomics tools for image analyses and clinical prediction in nasopharyngeal carcinoma. Br J Radiol. 2019;92(1102):20190271.
Bogowicz M, Leijenaar RTH, Tanadini-Lang S, Riesterer O, Pruschy M, Studer G, et al. Post-radiochemotherapy PET radiomics in head and neck cancer—The influence of radiomics implementation on the reproducibility of local control tumor models. Radiother Oncol. 2017;125(3):385–91.
Im HJ, Bradshaw T, Solaiyappan M, Cho SY. Current methods to define metabolic tumor volume in positron emission tomography: which one is better? Nucl Med Mol Imaging. 2018;52(1):5–15.
Song F, Guo Z, Mei D. Feature selection using principal component analysis. International conference on system science, engineering design and manufacturing informatization, Yichang, 2010. p. 27–30.
Lu L, Sun SH, Yang H, Guo P, Schwartz LH, et al. Radiomics prediction of EGFR status in lung cancer-our experience in using multiple feature extractors and the cancer imaging archive data. Tomography. 2020;6(2):223–30.
Fornacon-Wood I, Mistry H, Ackermann CJ, Blackhall F, McPartlin A, Faivre-Finn C, et al. Reliability and prognostic value of radiomic features are highly dependent on choice of feature extraction platform. Eur Radiol. 2020;30(11):6241–50.
Foy JJ, Armato SG, Al-Hallaq H. Effects of variability in radiomics software packages on classifying patients with radiation pneumonitis. J Med Imaging. 2020;7(1):014504.
Author information
Authors and Affiliations
Contributions
ML and RM conceptualized the paper; ML and RM evaluated and reported the imaging findings; RM carried out the statistical analysis; RS provided medical advice; ML, RM, and RS drafted the manuscript; all the authors revised the paper and approved its final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
All the procedures performed in this study involving human participants were in accordance with international ethical standards detailed in the 1964 Declaration of Helsinki and its later amendments, and according to the Italian Personal Data Protection Code for scientific research.
Informed consent
Nothing to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Larobina, M., Megna, R. & Solla, R. Comparison of three freeware software packages for 18F-FDG PET texture feature calculation. Jpn J Radiol 39, 710–719 (2021). https://doi.org/10.1007/s11604-021-01100-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11604-021-01100-0