Abstract
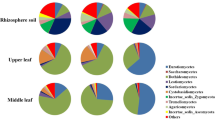
Multiple biotic and abiotic parameters influence the dynamics of individual fungal species and entire communities. Major drivers for tropical plant endophytes are undoubtedly seasonality, local habitat conditions and biogeography. However, host specialization and tissue preferences also contribute to the structuring of endophytic mycobiomes. To elucidate such specializations and preferences, we sampled two commercially important, unrelated plant species, Amorphophallus albispathus and Camellia sinensis (tea plant) simultaneously at close proximity. The mycobiomes of different tissue types were assessed with high-throughput amplicon sequencing of the internal transcribed spacer DNA region. Both plants hosted different fungal communities and varied in α- and β-diversity, despite their neighboring occurrence. However, the fungal assemblages of Amorphophallus leaflets shared taxa with the mycobiomes of tea leaves, thereby suggesting common driving forces for leaf-inhabiting fungi irrespective of host plant identity. The mycobiome composition and diversity of tea leaves was clearly driven by leaf age. We suggest that the very youngest tea leaves are colonized by stochastic processes, while mycobiomes of old leaves are rather similar as the result of progressive succession. The biodiversity of fungi associated with A. albispathus was characterized by a large number of unclassified OTUs (at genus and species level) and by tissue-specific composition.This study is the first cultivation-independent high-throughput assessment of fungal biodiversity of an Amorphophallus species, and additionally expands the knowledge base on fungi associated with tea plants.





Similar content being viewed by others
References
Abarenkov K, Adams RI, Laszlo I et al (2016) Annotating public fungal ITS sequences from the built environment according to the MIxS-built environment standard – a report from a may 23-24, 2016 workshop (Gothenburg, Sweden). MycoKeys 16:1–15
Agusta A, Ohashi K, Shibuya H (2006) Composition of the endophytic filamentous fungi isolated from the tea plant Camellia sinensis. J Nat Med 60:268–272
Arnold AE, Lutzoni F (2007) Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88:541–549
Bálint M, Schmidt P-A, Sharma R, Thines M, Schmitt I (2014) An Illumina metabarcoding pipeline for fungi. Ecol Evol 4:2642–2653
Barthlott W, Szarzynski J, Vlek P, Lobin W, Korotkova N (2009) A torch in the rain forest: thermogenesis of the titan arum (Amorphophallus titanum). Plant Biol 11:499–505
Beath DDN (1996) Pollination of Amorphophallus johnsonii (Araceae) by carrion beetles (Phaeochrous amplus) in a Ghanaian rain forest. J Trop Ecol 12:409
Beenken L, Senn-Irlet B (2016) Neomycetes in Switzerland – state of knowledge and estimation of potential risks of alien fungi associated with plant. WSL Berichte 50, 50p., Eidg. Forschungsanstalt WSL, Birmensdorf, Switzerland
Bengtsson-Palme J, Ryberg M, Hartmann M et al (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919
Brossa R, Casals I, Pintó-Marijuan M, Fleck I (2009) Leaf flavonoid content in Quercus ilex L. resprouts and its seasonal variation. Trees 23:401–408
Brown SP, Veach AM, Rigdon-Huss AR et al (2015) Scraping the bottom of the barrel: are rare high throughput sequences artifacts? Fungal Ecol 13:221–225
Brundrett MC (2006) Diversity and classification of mychorrhizal and endophytic fungi. In: Schulz BJE, Boyle CJC, Sieber TN (eds) Microbial root endophytes. Springer, Berlin, pp 281–298
Cabrera C, Artacho R, Giménez R (2006) Beneficial effects of green tea—a review. J am Coll. Nutrition 25:79–99
Carroll G, Petrini O (1983) Patterns of substrate utilization by some fungal endophytes from coniferous foliage. Mycologia 75:53
Chen J-L, Lin Y-R, Hou C-L, Wang S-J (2012) Species of Rhytismataceae on Camellia spp. from the Chinese mainland. Mycotaxon 118:219–230
Chen KL, Kirschner R (2017) Fungi from leaves of lotus (Nelumbo nucifera). Mycol Prog. https://doi.org/10.1007/s11557-017-1324-y
Chua M, Baldwin TC, Hocking TJ, Chan K (2010) Traditional uses and potential health benefits of Amorphophallus konjac K. Koch ex N.E.Br. J Ethnopharmacol 128:268–278
Collado J, Platas G, Pelaez F (1996) Fungal endophytes in leaves, twigs and bark of Quercus ilex from Central Spain. Nova Hedwigia 63:347–360
Collado J, Platas G, Gonzales I, Pelaez F (1999) Geographical and seasonal influences on the distribution of fungal endophytes in Quercus ilex. New Phytol 144:525–532
Cusimano N, Bogner J, Mayo SJ, Boyce PC, Wong SY, Hesse M, Hetterscheid WLA, Keating RC, French JC (2011) Relationships within the Araceae: comparison of morphological patterns with molecular phylogenies. Am J Bot 98:654–668
Daglia M, Antiochia R, Sobolev AP, Mannina L (2014) Untargeted and targeted methodologies in the study of tea (Camellia sinensis L.) Food Res Int 63:275–289
Dean RA, Talbot NJ, Ebbole DJ et al (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986
Delaye L, García-Guzmán G, Heil M (2013) Endophytes versus biotrophic and necrotrophic pathogens—are fungal lifestyles evolutionarily stable traits? Fungal Divers 60:125–135
Deng Y, Zhu Y, Wang P et al (2011) Complete genome sequence of Bacillus subtilis BSn5, an endophytic bacterium of Amorphophallus konjac with antimicrobial activity for the plant pathogen Erwinia carotovora subsp. carotovora. J Bacteriol 193:2070–2071
Doilom M, Dissanayake AJ, Wanasinghe DN et al (2016) Microfungi on Tectona grandis (teak) in northern Thailand. Fungal Divers. https://doi.org/10.1007/s13225-016-0368-7
Eberlein C, Heuer H, Vidal S, Westphal A (2016) Microbial communities in Globodera pallida females raised in potato monoculture soil. Phytopathology 106:581–590
Eusemann P, Schnittler M, Nilsson RH et al (2016) Habitat conditions and phenological tree traits overrule the influence of tree genotype in the needle mycobiome- Picea glauca system at an arctic treeline ecotone. New Phytol. https://doi.org/10.1111/nph.13988
Fang W, Yang L, Zhu X, Zeng L, Li X (2013) Seasonal and habitat dependent variations in culturable endophytes of Camellia sinensis. J Plant Pathol Microbiol 4:3. https://doi.org/10.4172/2157-7471.1000169
Fisher PJ, Anson AE, Petrini O (1986) Fungal endophytes in Ulex europaeus and Ulex gallii. Trans Br Mycol Soc 86:153–156
Friedman M (2007) Overview of antibacterial, antitoxin, antiviral, and antifungal activities of tea flavonoids and teas. Mol Nutri Food Res 51:116–134
Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A (2012) The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 25:106–141
Gille S, Cheng K, Skinner ME et al (2011) Deep sequencing of voodoo lily (Amorphophallus konjac): an approach to identify relevant genes involved in the synthesis of the hemicellulose glucomannan. Planta 234:515–526
Graham HN (1992) Green tea composition, consumption, and polyphenol chemistry. Prev Med 21:334–350
Hane JK, Lowe RGT, Solomon PS et al (2007) Dothideomycete plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell Online 19:3347–3368
Harbowy ME, Balentine DA, Davies AP, Cai Y (1997) Tea chemistry. Crit Rev Plant Sci 16:415–480
He Y, Caporaso JG, Jiang X-T et al (2015) Stability of operational taxonomic units: an important but neglected property for analyzing microbial diversity. Microbiome 3:20. https://doi.org/10.1186/s40168-015-0081-x
Hejnowicz Z, Barthlott W (2005) Structural and mechanical peculiarities of the petioles of giant leaves of Amorphophallus (Araceae). Am J Bot 92:391–403
Hetterscheid WLA (1994) Notes on the genus Amorphophallus (Araceae) – 2. Blumea 39:237–281
Higgins KL, Coley PD, Kursar TA, Arnold AE (2011) Culturing and direct PCR suggest prevalent host generalism among diverse fungal endophytes of tropical forest grasses. Mycologia 103:247–260
Hill MO (1973) Diversity and evenness: a unifying notation and its consequences. Ecology 54:427–432
Hunter PJ, Hand P, Pink D, Whipps JM, Bending GD (2010) Both leaf properties and microbe-microbe interactions influence within-species variation in bacterial population diversity and structure in the lettuce (Lactuca species) phyllosphere. Appl Environ Microbiol 76:8117–8125
Islam MT, Croll D, Gladieux P et al (2016) Emergence of wheat blast in Bangladesh was caused by a south American lineage of Magnaporthe oryzae. BMC Biol 14:84
Jensen AM, Warren JM, Hanson PJ, Childs J, Wullschleger SD (2015) Needle age and season influence photosynthetic temperature response and total annual carbon uptake in mature Picea mariana trees. Ann Bot 116:821–832
Johnston PR, Sutherland PW, Joshee S (2006) Visualising endophytic fungi within leaves by detection of (1′3)-β-d-glucans in fungal cell walls. Mycologist 20:159–162
Joshee S, Paulus BC, Park D, Johnston PR (2009) Diversity and distribution of fungal foliar endophytes in New Zealand Podocarpaceae. Mycol Res 113:1003–1015
Jumpponen A, Jones K (2009) Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperate Quercus macrocarpa phyllosphere. New Phytol 184:438–448
Jumpponen A, Jones K (2010) Seasonally dynamic fungal communities in the Quercus macrocarpa phyllosphere differ between urban and nonurban environments. New Phytol 186:496–513
Kembel SW, Mueller RC (2014) Plant traits and taxonomy drive host associations in tropical phyllosphere fungal communities. Botany 92:303–311
Khan A, Rahman M, Islam M (2008) Antibacterial, antifungal and cytotoxic activities of amblyone isolated from Amorphophallus campanulatus. Indian J Pharm 40:41
Kirschner R, Hou C-L, Chen C-J (2009) Co-occurrence of Pseudocercospora species and rhytismatalean ascomycetes on maple and Camellia in Taiwan. Mycol Prog 8:1–8
Kite GC, Hetterschieid WLA (1997) Inflorescence odours of Amorphophallus and Pseudodracontium (Araceae). Phytochemistry 46:71–75
Koljalg U, Nilsson R, Abarenkov K et al (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277
Kowalski T, Holdenrieder O (2009) The teleomorph of Chalara fraxinea, the causal agent of ash dieback. For Pathol 39:304–308
Kusari P, Kusari S, Spiteller M, Kayser O (2013) Endophytic fungi harbored in Cannabis sativa L.: diversity and potential as biocontrol agents against host plant-specific phytopathogens. Fungal Divers 60:137–151
Langarica-Fuentes A, Handley PS, Houlden A, Fox G, Robson GD (2014) An investigation of the biodiversity of thermophilic and thermotolerant fungal species in composts using culture-based and molecular techniques. Fungal Ecol 11:132–144
Lee J-E, Lee B-J, Chung JO, Hwang J-A, Lee S-J, Lee C-H, Hong Y-S (2010) Geographical and climatic dependencies of green tea (Camellia sinensis) metabolites: a 1 H NMR-based metabolomics study. J Agric Food Chem 58:10582–10589
Lee J-E, Lee B-J, Hwang J et al (2011) Metabolic dependence of green tea on plucking positions revisited: a metabolomic study. J Agric Food Chem 59:10579–10585
Liu F, Weir BS, Damm U et al (2015) Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 35:63–86
Lodge DJ, Cantrell S (1995) Fungal communities in wet tropical forests: variation in time and space. Can J Bot 73:1391–1398
Matulich KL, Weihe C, Allison SD et al (2015) Temporal variation overshadows the response of leaf litter microbial communities to simulated global change. ISME J. https://doi.org/10.1038/ismej.2015.58
May RM (1991) A fondness for fungi. Nature 352:475–476
Misra RS, Sriram S, Nedunchezhiyan M, Mohandas C (2003) Field and storage diseases of Amorphophallus and their management. Aroideana 26:42–53
Müller T, Ruppel S (2014) Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol Ecol 87:2–17
Nasanit R, Tangwong-o-thai A, Tantirungkij M, Limtong S (2015) The assessment of epiphytic yeast diversity in sugarcane phyllosphere in Thailand by culture-independent method. Fungal Biol 119:1145–1157
Navas-Molina J, Peralta-Sanchez J, Gonzalez A et al (2013) Advancing our understanding of the human microbiome using QIIME. Methods Enzymol 531:371–444
Nilsson RH, Hyde KD, Pawłowska J et al (2014) Improving ITS sequence data for identification of plant pathogenic fungi. Fungal Divers 67:11–19
Olbrich M, Knappe C, Wenig M et al (2010) Ozone fumigation (twice ambient) reduces leaf infestation following natural and artificial inoculation by the endophytic fungus Apiognomonia errabunda of adult European beech trees. Environ Pollut 158:1043–1050
Osono T (2008) Endophytic and epiphytic phyllosphere fungi of Camellia japonica: seasonal and leaf age-dependent variations. Mycologia 100:387–391
Peršoh D (2013) Factors shaping community structure of endophytic fungi–evidence from the Pinus-Viscum-system. Fungal Divers 60:55–69
Peršoh D (2015) Plant-associated fungal communities in the light of meta’omics. Fungal Divers 75:1–25
Petrini O, Fisher PJ (1990) Occurrence of fungal endophytes in twigs of Salix fragilis and Quercus robur. Mycol Res 94:1077–1080
Piepenbring M, Hofmann TA, Miranda E, Cáceres O, Unterseher M (2015) Leaf shedding and weather in tropical dry-seasonal forest shape the phenology of fungi – lessons from two years of monthly surveys in southwestern Panama. Fungal Ecol 18:83–92
Pitkaranta M, Meklin T, Hyvarinen A et al (2008) Analysis of fungal flora in indoor dust by ribosomal DNA sequence analysis, quantitative PCR, and culture. Appl Environ Microbiol 74:233–244
Punekar SA, Kumaran KPN (2010) Pollen morphology and pollination ecology of Amorphophallus species from north western Ghats and Konkan region of India. Flora 205:326–336
Rodriguez R, White JJ, Arnold A, Redman R (2009) Fungal endophytes: diversity and functional roles. New Phytol 182:314–330
Sansone C, Rita Massardo D, Pontieri P et al (2007) Isolation of a psychrotolerant Debaryomyces hansenii strain from fermented tea plant (Camellia sinensis) leaves. J Plant Interact 2:169–174
Siddique AB, Unterseher M (2016) A cost-effective and efficient strategy for Illumina sequencing of fungal communities: a case study of beech endophytes identified elevation as main explanatory factor for diversity and community composition. Fungal Ecol 20:175–185
Siddique AB, Khokon AM, Unterseher M (2017) What do we learn from cultures in the omics age? High-throughput sequencing and cultivation of leaf-inhabiting endophytes from beech (Fagus sylvatica L.) revealed complementary community composition but similar correlations with local habitat conditions. MycoKeys 20:1–16
Smith FA, Smith SE (1997) Tansley review no. 96. Structural diversity in (vesicular)-arbuscular mycorrhizal symbioses. New Phytol 137:373–388
Solis MJL, Dela Cruz TE, Schnittler M, Unterseher M (2016) The diverse community of leaf-inhabiting fungal endophytes from Philippine natural forests reflects phylogenetic patterns of their host plant species Ficus benjamina, F. elastica and F. religiosa. Mycoscience. https://doi.org/10.1016/j.myc.2015.10.002
Song R, Kelman D, Johns KL, Wright AD (2012) Correlation between leaf age, shade levels, and characteristic beneficial natural constituents of tea (Camellia sinensis) grown in Hawaii. Food Chem 133:707–714
Stajich JE, Berbee ML, Blackwell M et al (2009) The fungi. Curr Biol 19:R840–R845
Stone JK (1988) Fine structure of latent infections by on Douglas-fir, with observations on uninfected epidermal cells. Can J Bot 66:45–54
Suryanarayanan TS, Murali TS, Thirunavukkarasu N et al (2011) Endophytic fungal communities in woody perennials of three tropical forest types of the western Ghats, southern India. Biodivers Conserv 20:913–928
Tondello A, Vendramin E, Villani M, Baldan B, Squartini A (2012) Fungi associated with the southern Eurasian orchid Spiranthes spiralis (L.) Chevall. Fungal Biol 116:543–549
Unterseher M, Reiher A, Finstermeier K, Otto P, Morawetz W (2007) Species richness and distribution patterns of leaf-inhabiting endophytic fungi in a temperate forest canopy. Mycol Prog 6:201–212
Unterseher M, Gazis R, Chaverri P, Guarniz CFG, Tenorio DHZ (2013) Endophytic fungi from Peruvian highland and lowland habitats form distinctive and host plant-specific assemblages. Biodivers Conserv 22:999–1016
Unterseher M, Siddique AB, Brachmann A, Peršoh D (2016) Diversity and composition of the leaf mycobiome of beech (Fagus sylvatica) are affected by local habitat conditions and leaf biochemistry. PLoS ONE 11:e0152878
Viret O, Petrini O (1994) Colonization of beech leaves (Fagus sylvatica) by the endophyte Discula umbrinella (teleomorph: Apiognomonia errabunda). Mycol Res 98:423–432
Wiseman SA, Balentine DA, Frei B (1997) Antioxidants in tea. Crit Rev Food Sci Nutri 37:705–718
Xu A, Wang Y, Wen J et al (2011) Fungal community associated with fermentation and storage of Fuzhuan brick-tea. Int J Food Microbiol 146:14–22
Yang CS, Wang H, Li GX et al (2011) Cancer prevention by tea: evidence from laboratory studies. Pharmacol Res 64:113–122
Yu L, Zhao JR, SG X et al (2014) First report of gray mold on Amorphophallus muelleri caused by Botrytis cinerea in China. Plant Dis 98:692–692
Zambell CB, White JF (2015) In the forest vine Smilax rotundifolia, fungal epiphytes show site-wide spatial correlation, while endophytes show evidence of niche partitioning. Fungal Divers 75:279–297
Zheng X, Pan C, Diao Y et al (2013) Development of microsatellite markers by transcriptome sequencing in two species of Amorphophallus (Araceae). BMC Genomics 14:490
Zheng W-J, Wan X-C, Bao G-H (2015) Brick dark tea: a review of the manufacture, chemical constituents and bioconversion of the major chemical components during fermentation. Phytochem Rev 14:499–523
Acknowledgements
UM greatly acknowledges financial support from the German Science Foundation (DFG) under grant number UN 262/9-1, the German Academic Exchange Service (DAAD) for a travel grant to Northern Thailand and the Mushroom Research Centre. WC acknowledges financial support from the Marie Sklodowska-Curie action CRYPTRANS. UM also thanks Mushroom Research Foundation for the support of the field work at Mushroom Research Center (MRC), Chiang Mai. Klaus Fischer (Greifswald) and the DFG Research Training Group GRK 2010 RESPONSE are acknowledged for support of a PhD student workshop about metabarcoding during which most data were analyzed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Dominik Begerow
Rights and permissions
About this article
Cite this article
Unterseher, M., Karunarathna, S.C., Cruz, G.R. et al. Mycobiomes of sympatric Amorphophallus albispathus (Araceae) and Camellia sinensis (Theaceae) – a case study reveals clear tissue preferences and differences in diversity and composition. Mycol Progress 17, 489–500 (2018). https://doi.org/10.1007/s11557-018-1375-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-018-1375-8