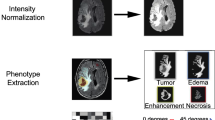
Abstract
Purpose
Low-grade gliomas (LGG) are classified into three distinct groups based on their IDH1 mutation and 1p/19q codeletion status, each of which is associated with a different clinical expression. The genomic sub-classification of LGG requires tumor sampling via neurosurgical procedures. The aim of this study was to evaluate the radiomics approach for noninvasive classification of patients with LGG and IDH mutation, based on their 1p/19q codeletion status, by testing different classifiers and assessing the contribution of the different MR contrasts.
Methods
Preoperative MRI scans of 47 patients diagnosed with LGG with IDH1-mutated tumors and a genetic analysis for 1p/19q deletion status were included in this study. A total of 152 features, including size, location and texture, were extracted from fluid-attenuated inversion recovery images, \(\hbox {T}_{2}\)-weighted images (WI) and post-contrast \(\hbox {T}_{1}\hbox {WI}\). Classification was performed using 17 machine learning classifiers. Results were evaluated by a fivefold cross-validation analysis.
Results
Radiomic analysis differentiated tumors with 1p/19q intact (\(n=21\); astrocytomas) from those with 1p/19q codeleted (\(n=26\); oligodendrogliomas). Best classification was obtained using the Ensemble Bagged Trees classifier, with sensitivity \(=\) 92%, specificity \(=\) 83% and accuracy \(=\) 87%, and with area under the curve \(=\) 0.87. Tumors with 1p/19q intact were larger than those with 1p/19q codeleted (\(46.2\pm 30.0\) vs. \(30.8\pm 16.8\) cc, respectively; \(p=0.03\)) and predominantly located to the left insula (\(p=0.04\)).
Conclusion
The proposed method yielded good discrimination between LGG with and without 1p/19q codeletion. Results from this study demonstrate the great potential of this method to aid decision-making in the clinical management of patients with LGG.



Similar content being viewed by others

References
Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, Robertson AG, Noushmehr H, Laird PW, Cherniack AD, Akbani R, Huse JT, Ciriello G, Poisson LM, Barnholtz-Sloan JS, Berger MS, Brennan C, Colen RR, Colman H, Flanders AE, Giannini C, Grifford M, Iavarone A, Jain R, Joseph I, Kim J, Kasaian K, Mikkelsen T, Murray BA, O’Neill BP, Pachter L, Parsons DW, Sougnez C, Sulman EP, Vandenberg SR, Van Meir EG, von Deimling A, Zhang H, Crain D, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton T, Sherman M, Yena P, Black A, Bowen J, Dicostanzo K, Gastier-Foster J, Leraas KM, Lichtenberg TM, Pierson CR, Ramirez NC, Taylor C, Weaver S, Wise L, Zmuda E, Davidsen T, Demchok JA, Eley G, Ferguson ML, Hutter CM, Mills Shaw KR, Ozenberger BA, Sheth M, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Ayala B, Baboud J, Chudamani S, Jensen MA, Liu J, Pihl T, Raman R, Wan Y, Wu Y, Ally A, Auman JT, Balasundaram M, Balu S, Baylin SB, Beroukhim R, Bootwalla MS, Bowlby R, Bristow CA, Brooks D, Butterfield Y, Carlsen R, Carter S, Chin L, Chu A, Chuah E, Cibulskis K, Clarke A, Coetzee SG, Dhalla N, Fennell T, Fisher S, Gabriel S, Getz G, Gibbs R, Guin R, Hadjipanayis A, Hayes DN, Hinoue T, Hoadley K, Holt RA, Hoyle AP, Jefferys SR, Jones S, Jones CD, Kucherlapati R, Lai PH, Lander E, Lee S, Lichtenstein L, Ma Y, Maglinte DT, Mahadeshwar HS, Marra MA, Mayo M, Meng S, Meyerson ML, Mieczkowski PA, Moore RA, Mose LE, Mungall AJ, Pantazi A, Parfenov M, Park PJ, Parker JS, Perou CM, Protopopov A, Ren X, Roach J, Sabedot TS, Schein J, Schumacher SE, Seidman JG, Seth S, Shen H, Simons JV, Sipahimalani P, Soloway MG, Song X, Sun H, Tabak B, Tam A, Tan D, Tang J, Thiessen N, Triche T Jr, Van Den Berg DJ, Veluvolu U, Waring S, Weisenberger DJ, Wilkerson MD, Wong T, Wu J, Xi L, Xu AW, Yang L, Zack TI, Zhang J, Aksoy BA, Arachchi H, Benz C, Bernard B, Carlin D, Cho J, DiCara D, Frazer S, Fuller GN, Gao J, Gehlenborg N, Haussler D, Heiman DI, Iype L, Jacobsen A, Ju Z, Katzman S, Kim H, Knijnenburg T, Kreisberg RB, Lawrence MS, Lee W, Leinonen K, Lin P, Ling S, Liu W, Liu Y, Liu Y, Lu Y, Mills G, Ng S, Noble MS, Paull E, Rao A, Reynolds S, Saksena G, Sanborn Z, Sander C, Schultz N, Senbabaoglu Y, Shen R, Shmulevich I, Sinha R, Stuart J, Sumer SO, Sun Y, Tasman N, Taylor BS, Voet D, Weinhold N, Weinstein JN, Yang D, Yoshihara K, Zheng S, Zhang W, Zou L, Abel T, Sadeghi S, Cohen ML, Eschbacher J, Hattab EM, Raghunathan A, Schniederjan MJ, Aziz D, Barnett G, Barrett W, Bigner DD, Boice L, Brewer C, Calatozzolo C, Campos B, Carlotti CG Jr, Chan TA, Cuppini L, Curley E, Cuzzubbo S, Devine K, DiMeco F, Duell R, Elder JB, Fehrenbach A, Finocchiaro G, Friedman W, Fulop J, Gardner J, Hermes B, Herold-Mende C, Jungk C, Kendler A, Lehman NL, Lipp E, Liu O, Mandt R, McGraw M, McLendon R, McPherson C, Neder L, Nguyen P, Noss A, Nunziata R, Ostrom QT, Palmer C, Perin A, Pollo B, Potapov A, Potapova O, Rathmell WK, Rotin D, Scarpace L, Schilero C, Senecal K, Shimmel K, Shurkhay V, Sifri S, Singh R, Sloan AE, Smolenski K, Staugaitis SM, Steele R, Thorne L, Tirapelli DP, Unterberg A, Vallurupalli M, Wang Y, Warnick R, Williams F, Wolinsky Y, Bell S, Rosenberg M, Stewart C, Huang F, Grimsby JL, Radenbaugh AJ, Zhang J (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372(26):2481–2498
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321(5897):1807–12
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360(8):765–773
Cohen AL, Holmen SL, Colman H (2013) IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep 13(5):345
Boots-Sprenger SH, Sijben A, Rijntjes J, Tops BB, Idema AJ, Rivera AL, Bleeker FE, Gijtenbeek AM, Diefes K, Heathcock L, Aldape KD, Jeuken JW, Wesseling P (2013) Significance of complete 1p/19q co-deletion, IDH1 mutation and MGMT promoter methylation in gliomas: use with caution. Mod Pathol 26(7):922–9
Chamberlain MC, Born D (2015) Prognostic significance of relative 1p/19q codeletion in oligodendroglial tumors. J Neuro Oncol 125(2):249–251
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Macarena I, Young RJ, Rubel J, Rosenblum M, Tisnado J, Briggs S, Arevalo-Perez J, Cross JR, Campos C, Straley K (2016) Integration of 2-hydroxyglutarate-proton magnetic resonance spectroscopy into clinical practice for disease monitoring in isocitrate dehydrogenase-mutant glioma. Neuro Oncology 18(2):283–290
Pope WB, Prins RM, Thomas MA, Nagarajan R, Yen KE, Bittinger MA, Salamon N, Chou AP, Yong WH, Soto H (2012) Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neuro Oncol 107(1):197–205
Andronesi OC, Rapalino O, Gerstner E, Chi A, Batchelor TT, Cahill DP, Sorensen AG, Rosen BR (2013) Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J Clin Investig 123(9):3659–3663
Lee S, Choi SH, Ryoo I, Yoon TJ, Kim TM, Lee S-H, Park C-K, Kim J-H, Sohn C-H, Park S-H (2015) Evaluation of the microenvironmental heterogeneity in high-grade gliomas with IDH1/2 gene mutation using histogram analysis of diffusion-weighted imaging and dynamic-susceptibility contrast perfusion imaging. J Neuro Oncol 121(1):141–150
Yamashita K, Hiwatashi A, Togao O, Kikuchi K, Hatae R, Yoshimoto K, Mizoguchi M, Suzuki S, Yoshiura T, Honda H (2016) MR imaging-based analysis of glioblastoma multiforme: estimation of IDH1 mutation status. Am J Neuroradiol 37(1):58–65
Yu J, Shi Z, Lian Y, Li Z, Liu T, Gao Y, Wang Y, Chen L, Mao Y (2017) Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol 27(8):3509–3522
Zhang B, Chang K, Ramkissoon S, Tanguturi S, Bi WL, Reardon DA, Ligon KL, Alexander BM, Wen PY, Huang RY (2017) Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncology 19(1):109–117
Yu J, Shi Z, Ji C, Lian Y, Wang Y, Chen L, Mao Y (2016) Anatomical location differences between mutated and wild-type isocitrate dehydrogenase 1 in low-grade gliomas. Int J Neurosci 2017:1–8
Akkus Z, Ali I, Sedlar J, Kline TL, Agrawal JP, Parney IF, Giannini C, Erickson BJ (2016) Predicting 1p19q chromosomal deletion of low-grade gliomas from MR images using deep learning. arXiv preprint arXiv:1611.06939
Zhou H, Vallières M, Bai HX, Su C, Tang H, Oldridge D, Zhang Z, Xiao B, Liao W, Tao Y (2017) MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncology 19(6):862–870
Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 3(5):4006
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48(4):441–446
Gillies RJ, Kinahan PE, Hricak H (2015) Radiomics: images are more than pictures, they are data. Radiology 278(2):563–577
Kotrotsou A, Zinn PO, Colen RR (2016) Radiomics in brain tumors: an emerging technique for characterization of tumor environment. Magn Reson Imaging Clin N Am 24(4):719–729
Lavon I, Refael M, Zelikovitch B, Shalom E, Siegal T (2010) Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro Oncology 12(2):173–180
Jenkinson M, Smith S (2001) A global optimisation method for robust affine registration of brain images. Med Image Anal 5(2):143–156
Smith SM (2002) Fast robust automated brain extraction. Hum Brain Mapp 17(3):143–155
Zhang Y, Brady M, Smith S (2001) Segmentation of brain MR images through a hidden Markov random field model and the expectation–maximization algorithm. IEEE Trans Med Imaging 20(1):45–57
Haralick RM, Shanmugam K (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 6:610–621
Kickingereder P, Bonekamp D, Nowosielski M, Kratz A, Sill M, Burth S, Wick A, Eidel O, Schlemmer HP, Radbruch A, Debus J, Herold-Mende C, Unterberg A, Jones D, Pfister S, Wick W, von Deimling A, Bendszus M, Capper D (2016) Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology 281(3):907–918
Kassner A, Thornhill RE (2010) Texture analysis: a review of neurologic MR imaging applications. AJNR Am J Neuroradiol 31(5):809–816
Skogen K, Schulz A, Dormagen JB, Ganeshan B, Helseth E, Server A (2016) Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol 85(4):824–829
Chang EF, Smith JS, Chang SM, Lamborn KR, Prados MD, Butowski N, Barbaro NM, Parsa AT, Berger MS, Mcdermott MM (2008) Preoperative prognostic classification system for hemispheric low-grade gliomas in adults. J Neurosurg 109(5):817–824
Zlatescu MC, TehraniYazdi A, Sasaki H, Megyesi JF, Betensky RA, Louis DN, Cairncross JG (2001) Tumor location and growth pattern correlate with genetic signature in oligodendroglial neoplasms. Cancer Res 61(18):6713–6715
Acknowledgements
To Esther Eshkol for editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. For this type of study, formal consent was not required.
Rights and permissions
About this article
Cite this article
Shofty, B., Artzi, M., Ben Bashat, D. et al. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int J CARS 13, 563–571 (2018). https://doi.org/10.1007/s11548-017-1691-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-017-1691-5