Abstract
Phylogenetic networks are generalizations of phylogenetic trees that allow the representation of reticulation events such as horizontal gene transfer or hybridization, and can also represent uncertainty in inference. A subclass of these, tree-based phylogenetic networks, have been introduced to capture the extent to which reticulate evolution nevertheless broadly follows tree-like patterns. Several important operations that change a general phylogenetic network have been developed in recent years and are important for allowing algorithms to move around spaces of networks; a vital ingredient in finding an optimal network given some biological data. A key such operation is the nearest neighbour interchange, or NNI. While it is already known that the space of unrooted phylogenetic networks is connected under NNI, it has been unclear whether this also holds for the subspace of tree-based networks. In this paper, we show that the space of unrooted tree-based phylogenetic networks is indeed connected under the NNI operation. We do so by explicitly showing how to get from one such network to another one without losing tree-basedness along the way. Moreover, we introduce some new concepts, for instance “shoat networks”, and derive some interesting aspects concerning tree-basedness. Last, we use our results to derive an upper bound on the size of the space of tree-based networks.







Similar content being viewed by others
Notes
Because of their close resemblance to the juvenile boars that frequent the streets of northern Germany.
Note that a binary phylogenetic tree with n leaves has precisely \(2n-2\) vertices (Semple and Steel 2003). Any of the extra k edges added to such a tree will induce two new vertices. In total, this is \(2n-2+2k = 2(n+k-1)\) vertices.
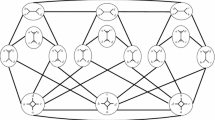
We used the computer algebra system Mathematica (Wolfram 2017) to verify that the shortest path from network (i) in Fig. 7 to a network isomorphic to (ii) requires 5 NNI moves. While this is tricky to see, it is combinatorially rather easy to see that network (i) has 24 1-step NNI neighbours, which can be divided into two classes (i.e. the NNI neighbourhood of network (i) contains only two non-isomorphic networks): those isomorphic to (i), and those isomorphic to a specific different network, which is in fact tree-based. So network (ii) cannot be in the 1-step neighbourhood of (i).
References
Allen BL, Steel M (2001) Subtree transfer operations and their induced metrics on evolutionary trees. Ann Combin 5(1):1–15
Fischer M, Francis A (2020) How tree-based is network? Proximity measures for unrooted phylogenetic networks. Dis Appl Math. https://doi.org/10.1016/j.dam.2019.12.019
Fischer M, Galla M, Herbst L, Long Y, Wicke K (2018) Unrooted non-binary tree-based phylogenetic networks. arXiv:1810.06853
Fischer M, Galla M, Herbst L, Long Y, Wicke K (2020) Classes of tree-based networks. In: press at Visual Computing for Industry, Biomedicine, and Art, Special issue on Applications of Graph Theory in Bioinformatics
Francis AR, Steel M (2015) Which phylogenetic networks are merely trees with additional arcs? Syst Biol 64(5):768–777
Francis A, Huber KT, Moulton V (2018) Tree-based unrooted phylogenetic networks. Bull Math Biol 80(2):404–416
Francis A, Huber KT, Moulton V, Wu T (2018) Bounds for phylogenetic network space metrics. J Math Biol 76(5):1229–1248
Francis A, Semple C, Steel M (2018) New characterisations of tree-based networks and proximity measures. Adv Appl Math 93:93–107
Gambette P, Berry V, Paul C (2012) Quartets and unrooted phylogenetic networks. J Bioinf Comput Biol 10(04):1250004
Gambette P, van Iersel L, Jones M, Lafond M, Pardi F, Scornavacca C (2017) Rearrangement moves on rooted phylogenetic networks. PLOS Comput Biol 13(8):e1005611
Hendriksen M (2018) Tree-based unrooted nonbinary phylogenetic networks. Math Biosci 302:131–138
Höhna S, Drummond A (2011) Guided tree topology proposals for Bayesian phylogenetic inference. Syst Biol 61(1):1–11
Huber KT, Moulton V, Wu T (2016) Transforming phylogenetic networks: moving beyond tree space. J Theor Biol 404:30–39
Huson DH, Rupp R, Scornavacca C (2010) Phylogenetic networks: concepts, algorithms and applications. Cambridge University Press, Cambridge
Janssen R, Klawitter J (2019) Rearrangement operations on unrooted phylogenetic networks. Theory Appl Graphs 6(2):6
Jetten L, van Iersel L (2016) Nonbinary tree-based phylogenetic networks. IEEE/ACM Transactions on Computational Biology and Bioinformatics
Lakner C, van der Mark P, Huelsenbeck JP, Larget B, Ronquist F (2008) Efficiency of Markov Chain Monte Carlo tree proposals in Bayesian phylogenetics. Syst Biol 57(1):86–103
Pons JC, Semple C, Steel M (2018) Tree-based networks: characterisations, metrics, and support trees. J Math Biol 78(4):899–918
Robinson DF (1971) Comparison of labeled trees with valency three. J Combin Theory Ser B 11(2):105–119
Semple C, Steel M (2003) Phylogenetics (Oxford lecture series in mathematics and its applications). Oxford University Press, Oxford
Steel M (2016) Phylogeny: discrete and random processes in evolution. SIAM, New Delhi
Whidden C, Matsen IV, Frederick A (2015) Quantifying MCMC exploration of phylogenetic tree space. Syst Biol 64(3):472–491
Wolfram Research, Inc. Mathematica, Version 10.3, (2017) Champaign, IL
Acknowledgements
MF wishes to thank the DAAD for conference travel funding to the Annual New Zealand Phylogenomics Meeting, where partial results leading to this manuscript were achieved.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fischer, M., Francis, A. The Space of Tree-Based Phylogenetic Networks. Bull Math Biol 82, 70 (2020). https://doi.org/10.1007/s11538-020-00744-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-020-00744-9