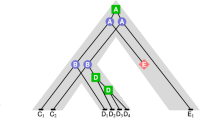
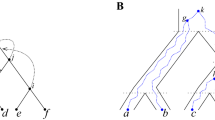
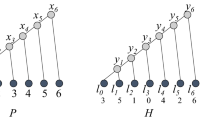
Abstract
An ancestral configuration is one of the combinatorially distinct sets of gene lineages that, for a given gene tree, can reach a given node of a specified species tree. Ancestral configurations have appeared in recursive algebraic computations of the conditional probability that a gene tree topology is produced under the multispecies coalescent model for a given species tree. For matching gene trees and species trees, we study the number of ancestral configurations, considered up to an equivalence relation introduced by Wu (Evolution 66:763–775, 2012) to reduce the complexity of the recursive probability computation. We examine the largest number of non-equivalent ancestral configurations possible for a given tree size n. Whereas the smallest number of non-equivalent ancestral configurations increases polynomially with n, we show that the largest number increases with \(k^n\), where k is a constant that satisfies \(\root 3 \of {3}\,\le \,k\,<\,1.503\). Under a uniform distribution on the set of binary labeled trees with a given size n, the mean number of non-equivalent ancestral configurations grows exponentially with n. The results refine an earlier analysis of the number of ancestral configurations considered without applying the equivalence relation, showing that use of the equivalence relation does not alter the exponential nature of the increase with tree size.








Similar content being viewed by others
References
Aho AV, Sloane NJA (1973) Some doubly exponential sequences. Fibonacci Q. 11:429–437
Allman ES, Degnan JH, Rhodes JA (2011) Identifying the rooted species tree from the distribution of unrooted gene trees under the coalescent. J Math Biol 62:833–862
Degnan JH, Salter LA (2005) Gene tree distributions under the coalescent process. Evolution 59:24–37
Disanto F, Rosenberg NA (2015) Coalescent histories for lodgepole species trees. J Comput Biol 22:918–929
Disanto F, Rosenberg NA (2016) Asymptotic properties of the number of matching coalescent histories for caterpillar-like families of species trees. IEEE/ACM Trans Comput Biol Bioinf 13:913–925
Disanto F, Rosenberg NA (2017) Enumeration of ancestral configurations for matching gene trees and species trees. J Comput Biol 24:831–850
Felsenstein J (1978) The number of evolutionary trees. Syst. Zool. 27:27–33
Felsenstein J (2004) Inferring phylogenies. Sinauer, Sunderland, MA
Flajolet P, Sedgewick R (2009) Analytic combinatorics. Cambridge University Press, Cambridge
Harding EF (1971) The probabilities of rooted tree-shapes generated by random bifurcation. Adv Appl Prob 3:44–77
Rosenberg NA (2006) The mean and variance of the numbers of \(r\)-pronged nodes and \(r\)-caterpillars in Yule-generated genealogical trees. Ann Comb 10:129–146
Rosenberg NA (2007) Counting coalescent histories. J Comput Biol 14:360–377
Rosenberg NA (2013) Coalescent histories for caterpillar-like families. IEEE/ACM Trans Comput Biol Bioinf 10:1253–1262
Rosenberg NA, Degnan JH (2010) Coalescent histories for discordant gene trees and species trees. Theor Pop Biol 77:145–151
Sedgewick R, Flajolet P (1996) An introduction to the analysis of algorithms. Addison-Wesley, Boston
Than C, Ruths D, Innan H, Nakhleh L (2007) Confounding factors in HGT detection: statistical error, coalescent effects, and multiple solutions. J Comput Biol 14:517–535
Wu Y (2012) Coalescent-based species tree inference from gene tree topologies under incomplete lineage sorting by maximum likelihood. Evolution 66:763–775
Acknowledgements
We thank Elizabeth Allman, James Degnan, and John Rhodes for discussions, and two reviewers for comments. Support was provided by National Institutes of Health grant R01 GM117590 and by a 2014 Rita Levi Montalcini grant to FD from the Ministero dell’Istruzione, dell’Università e della Ricerca.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1: Proof of (9)
Let \(C^*(r_S) = \{\gamma _{S,1}, \ldots , \gamma _{S,q} \}\) with \(c^*(r_S)=q\), and let \(C^*(r_L) = \{\gamma _{L,1}, \ldots ,\gamma _{L,Q} \}\), with \(c^*(r_L) = Q\). Because condition (8) is satisfied, the entire tree \(t_{r_S}\) can be displayed in \(t_{r_L}\), each configuration \(\gamma _{S,i} \in C^*(r_S)\) has exactly one corresponding configuration \(\gamma _{L,i} \in C^*(r_L)\) such that \(t_{r_S}(\gamma _{S,i}) \cong t_{r_L}(\gamma _{L,i})\), and \(Q\,\ge \,q\).
From (6), we obtain
which can be further decomposed as
We merge equivalent configurations to obtain \(C^*(r)\) from \(\tilde{C}(r)\). From (29), we remove those in \(\{\gamma _{S,1}, \ldots ,\gamma _{S,q} \} \otimes \{ \{r_L \} \} \), as they are equivalent to those in \(\{ \{ r_{S}\} \} \otimes \{\gamma _{L,1}, \ldots ,\gamma _{L,q} \}\). Thus, we take only q among the 2q configurations in (29). Moreover, due to the equivalence \(\gamma _{S,i} \cup \gamma _{L,j} \sim _r \gamma _{S,j} \cup \gamma _{L,i}\), we take only those configurations of the form \(\gamma _{S,i} \cup \gamma _{L,j}\) with \(i\,\le \,j\) among those in \(\{\gamma _{S,1}, \ldots ,\gamma _{S,q} \} \otimes \{\gamma _{L,1}, \ldots ,\gamma _{L,q} \}\). Thus, among the \(q^2\) configurations in (31)—those with \(1\,\le \,i, j\,\le \,q\)—we take only \(q(q+1)/2\) non-equivalent ones. No equivalences are possible among configurations in (28), (30), and (32), and all are retained in \(C^*(r)\). From (28)–(32), we then have
Replacing q by \(c^*(r_S)\) and Q by \(c^*(r_L)\) gives (9).
Appendix 2: Proof of (12)
The proof follows the approach of Aho and Sloane (1973, Sect. 3) for solving certain recurrences. From (11), we have \(x_{h+1} = x_h^2 [1 + 1/(2x_h) + 1/(2x_h^2) ]\). Taking the logarithm \(y_h = \log x_h\) yields \(y_{h+1} = 2y_h + \alpha _h\), where \(\alpha _h = \log [1+ {1}/{(2x_h)} + {1}/{(2x_h^2)}]\). Following Aho and Sloane (1973), \(y_h\) has solution
Converting back to \(x_h = \exp (y_h)\), from (33) we have
where the last step uses the fact that \(x_0=1/2\).
We then have
When \(h \rightarrow \infty \), the sum \(\sum _{i=h}^{\infty } 2^{h-i-1}\alpha _i\) converges to zero because it can be bounded \(0 \le \sum _{i=h}^{\infty } 2^{h-i-1}\alpha _i\,\le \,\alpha _h \sum _{i=h}^{\infty } 2^{h-i-1} = \alpha _h\), where because \(x_h \rightarrow \infty \) as \(h \rightarrow \infty \), \(\alpha _h \rightarrow 0\) as \(h \rightarrow \infty \). It follows that \(x_h/(k_0^*)^{(2^h)}\) converges to 1, producing (12).
Appendix 3: Properties of \(w'(n)\)
We prove that for each \(n\ge 2\), \(w'(n)\,\le \,n/2\), with equality only for \(n=2\), 4, or 6. The result is verified by direct computation of \(w'(n)\) for \(2\,\le \,n\,\le \,7\). For \(n\,\ge \,8\), by definition, \(w'(n)=\lfloor x \rfloor \), where x satisfies \(2^{x-2}+x=n-1\). Seeking a contradiction, suppose \(\lfloor x \rfloor = w'(n)\,\ge \,n/2\). Because \(x\,\ge \,\lfloor x \rfloor \), we would have \(x\,\ge \,n/2\), and therefore \(n-1=2^{x-2}+x\,\ge \,2^{n/2-2} + n/2 \ge 2(n/2 - 2) + n/2 = 3n/2-4\), noting that \(2^u\,\ge \,2u\) for \(u \ge 2\). The inequality \(n-1\,\ge \,3n/2-4\) cannot hold if \(n\,\ge \,8\). Therefore, when \(n\,\ge \,8\), we must have \(w'(n) < n/2\).
Appendix 4: Proof that Trees in \(T_{n,w}\) Satisfy (8) for \(w\,\ge \,2\)
We first prove that given any \(w\ge 2\), a caterpillar tree \(t_1\) of size \(|t_1| = w\) can be displayed in any tree \(t_2\) of size \(|t_2| \ge 2^{w-2}+1\) through a root configuration \(\gamma \) of \(t_2\), that is, \(t_1 \cong t_2(\gamma )\). The proof is by induction on w.
For \(w=2\), we have \(|t_2|\,\ge \,2\) and the result follows by taking the root configuration \(\gamma \) determined by the left and right descendants of the root in \(t_2\). For the inductive step, because \(|t_2|\,\ge \,2^{w-2}+1\), the larger root subtree of \(t_2\) has size at least \(\lceil |t_2|/2 \rceil \,\ge \,\lceil 2^{w-3}+1/2 \rceil = 2^{w-3} + 1 \). By the inductive hypothesis, the larger root subtree of \(t_2\) can display a caterpillar of size \(w-1\) through a root configuration \(\gamma '\). Taking the root configuration \(\gamma \) of \(t_2\) obtained as \(\gamma = \gamma ' \cup \{ \rho \}\), where \(\rho \) is the root of the smaller root subtree of \(t_2\), we have \(t_1 \cong t_2(\gamma )\) as desired.
Now suppose we are given a tree \(t \in T_{n,w}\), with \(2\,\le \,w \le w'(n)\). The smaller root subtree \(t_{r_S}\) of t is by definition a caterpillar of size \(w\,\ge \,2\), and the larger root subtree \(t_{r_L}\) has size \(|t_{r_L}| = n-w\). By definition, \(w\,\le \,w'(n) = \lfloor x \rfloor \,\le \,x\), where \(x = n - 2^{x-2} -1\), and therefore, \(w\,\le \,n - 2^{w-2} - 1\). In particular, \(|t_{r_L}| = n-w \ge 2^{w-2}+1\). From what we have shown above, a root configuration \(\gamma \) of \(t_{r_L}\) exists such that \(t_{r_S} \cong t_{r_L}(\gamma )\).
Appendix 5: Proof of (18)
Recall that for each tree \(t \in T_{n,w}\), the smaller root subtree \(t_{r_S}\) is a caterpillar of size \(w \in [1,w']\) and the larger root subtree \(t_{r_L}\) has size \(n-w\). Because we assume \(w < n/2\), \(t_{r_S}\) and \(t_{r_L}\) have different sizes and different unlabeled topologies. Given a tree \(\overline{t} \in T_{n-w}\), the number of trees in \(T_{n,w}\) such that \(t_{r_L} = \overline{t}\) (after rescaling labels for the taxa) is \({{n}\atopwithdelims (){w}} \gamma _w\), where \(\gamma _w\) is the number of caterpillar labeled topologies of size w. Dividing by \(|T_{n,w}| = {{n}\atopwithdelims (){w}} \gamma _w |T_{n-w}|\) yields the probability \(\mathbb {P}[t_{r_L}=\overline{t}|t \in T_{n,w}] = 1/|T_{n-w}|\) as desired.
Rights and permissions
About this article
Cite this article
Disanto, F., Rosenberg, N.A. On the Number of Non-equivalent Ancestral Configurations for Matching Gene Trees and Species Trees. Bull Math Biol 81, 384–407 (2019). https://doi.org/10.1007/s11538-017-0342-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-017-0342-x