Abstract
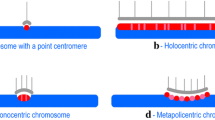
The centromere is a highly organized structure mainly composed of repeat sequences, which make this region extremely difficult for sequencing and other analyses. It plays a conserved role in equal division of chromosomes into daughter cells in both mitosis and meiosis. However, centromere sequences show notable plasticity. In a dicentric chromosome, one of the centromeres can become inactivated with the underlying DNA unchanged. Furthermore, formerly inactive centromeres can regain activity under certain conditions. In addition, neocentromeres without centromeric repeats have been found in a wide spectrum of species. This evidence indicates that epigenetic mechanisms together with centromeric sequences are associated with centromere specification.
Similar content being viewed by others
References
Alfenito MR, Birchler J A (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics, 135(2): 589–597
Allshire R C, Karpen G H (2008). Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet, 9(12): 923–937
Amor D J, Choo K H A (2002). Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet, 71(4): 695–714
Ananiev E V, Phillips R L, Rines H W (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc Natl Acad Sci U S A, 95(22): 13073–13078
Ando S, Yang H, Nozaki N, Okazaki T, Yoda K (2002). CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol, 22(7): 2229–2241
Birchler J A, Han F P (2009). Maize centromeres: structure, function, epigenetics. Annu Rev Genet, 43(1): 287–303
Blower M D, Sullivan B A, Karpen G H (2002). Conserved organization of centromeric chromatin in flies and humans. Dev Cell, 2(3): 319–330
Carlson W R (1969). Factors affecting preferential fertilization in maize. Genetics, 62(3): 543–554
Carlson W R, Phillips R (1986). The B-chromosome of maize. Crit Rev Plant Sci, 3(3): 201–226
Choo K H A (1997). Centromere DNA dynamics: latent centromeres and neocentromere formation. Am J Hum Genet, 61(6): 1225–1233
Dawe R K, Hiatt E N (2004). Plant neocentromeres: fast, focused, and driven. Chromosome Res, 12(6): 655–669
Dawe R K, Reed L M, Yu H G, Muszynski M G, Hiatt E N (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell, 11(7): 1227–1238
Guerra M, Cabral G, Cuacos M, González-García M, González-Sánchez M, Vega J, Puertas M J (2010). Neocentrics and holokinetics (holocentrics): chromosomes out of the centromeric rules. Cytogenet Genome Res, 129(1–3): 82–96
Hamant O, Golubovskaya I, Meeley R, Fiume E, Timofejeva L, Schleiffer A, Nasmyth K, Cande W Z (2005). A REC8-dependent plant Shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr Biol, 15(10): 948–954
Han F P, Gao Z, Birchler J A (2009). Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell, 21(7): 1929–1939
Han F P, Gao Z, Yu W C, Birchler J A (2007a). Minichromosome analysis of chromosome pairing, disjunction, and sister chromatid cohesion in maize. Plant Cell, 19(12): 3853–3863
Han F P, Lamb J C, Birchler J A (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci U S A, 103(9): 3238–3243
Han F P, Lamb J C, Yu W C, Gao Z, Birchler J A (2007b). Centromere function and nondisjunction are independent components of the maize B chromosome accumulation mechanism. Plant Cell, 19(2): 524–533
Henikoff S, Ahmad K, Malik H S (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science, 293(5532): 1098–1102
Higgins A W, Gustashaw K M, Willard H F (2005). Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res, 13(8): 745–762
Jiang J M, Birchler J A, Parrott W A, Dawe R K (2003). A molecular view of plant centromeres. Trends Plant Sci, 8(12): 570–575
Jin WW, Melo J R, Nagaki K, Talbert P B, Henikoff S, Dawe R K, Jiang J M (2004). Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell, 16(3): 571–581
Jones R N, Rees H (1982). B chromosomes. London, Academic Press
Lamb J C, Kato A, Birchler J A (2005). Sequences associated with A chromosome centromeres are present throughout the maize B chromosome. Chromosoma, 113(7): 337–349
Lin B Y (1978). Regional control of nondisjunction of the B chromosome in maize. Genetics, 90(3): 613–627
Malik H S, Henikoff S (2009). Major evolutionary transitions in centromere complexity. Cell, 138(6): 1067–1082
McClintock B (1939). The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci U S A, 25(8): 405–416
McClintock B (1941). The stability of broken ends of chromosomes in Zea mays. Genetics, 26(2): 234–282
Mroczek R J, Dawe R K (2003). Distribution of retroelements in centromeres and neocentromeres of maize. Genetics, 165(2): 809–819
Nagaki K, Song J Q, Stupar R M, Parokonny A S, Yuan Q P, Ouyang S, Liu J, Hsiao J, Jones K M, Dawe R K, Buell C R, Jiang J M (2003). Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics, 163(2): 759–770
Nasuda S, Hudakova S, Schubert I, Houben A, Endo T R (2005). Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci U S A, 102(28): 9842–9847
Page S L, Shaffer L G (1998). Chromosome stability is maintained by short intercentromeric distance in functionally dicentric human Robertsonian translocations. Chromosome Res, 6(2): 115–122
Presting G G, Malysheva L, Fuchs J, Schubert I Z (1998). A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J, 16(6): 721–728
Rhoades M M, Vilkomerson H (1942). On the anaphase movement of chromosomes. Proc Natl Acad Sci U S A, 28(10): 433–436
Roman H (1947). Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics, 32: 391–409
Roman H (1948). Directed fertilization in maize. Proc Natl Acad Sci USA, 34(2): 36–42
Shelby R D, Monier K, Sullivan K F (2000). Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol, 151(5): 1113–1118
Stimpson K M, Song I Y, Jauch A, Holtgreve-Grez H, Hayden K E, Bridger J M, Sullivan B A, Copenhaver G P (2010). Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet, 6(8): e1001061
Stimpson K M, Sullivan B A (2010). Epigenomics of centromere assembly and function. Curr Opin Cell Biol, 22(6): 1–9
Sullivan B A, Willard H F (1998). Stable dicentric X chromosomes with two functional centromeres. Nat Genet, 20(3): 227–228
Topp C N, Okagaki R J, Melo J R, Kynast R G, Phillips R L, Dawe R K (2009). Identification of a maize neocentromere in an oat-maize addition line. Cytogenet Genome Res, 124(3–4): 228–238
Van Hooser A A, Ouspenski I I, Gregson H C, Starr D A, Yen T J, Goldberg M L, Yokomori K, Earnshaw W C, Sullivan K F, Brinkley B R (2001). Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci, 114(Pt 19): 3529–3542
Voullaire L E, Slater H R, Petrovic V, Choo K H A (1993). A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet, 52(6): 1153–1163
Ward E J (1973). Nondisjunction: localization of the controlling site in the maize B chromosome. Genetics, 73(3): 387–391
Watanabe Y (2005). Shugoshin: guardian spirit at the centromere. Curr Opin Cell Biol, 17(6): 590–595
Yu W C, Han F P, Gao Z, Vega J M, Birchler J A (2007). Construction and behavior of engineered minichromosomes in maize. Proc Natl Acad Sci U S A, 104(21): 8924–8929
Yu W C, Lamb J C, Han F P, Birchler J A (2006). Telomere-mediated chromosomal truncation in maize. Proc Natl Acad Sci U S A, 103(46): 17331–17336
Zhang W L, Friebe B, Gill B S, Jiang J M (2010). Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma, 119(5): 553–563
Zheng Y Z, Roseman R R, Carlson WR (1999). Time course study of the chromosome-type breakage-fusion-bridge cycle in maize. Genetics, 153(3): 1435–1444
Zhong C X, Marshall J B, Topp C, Mroczek R, Kato A, Nagaki K, Birchler J A, Jiang J M, Dawe R K (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell, 14(11): 2825–2836
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Yin, W., Birchler, J.A. & Han, F. Maize centromeres: where sequence meets epigenetics. Front. Biol. 6, 102–108 (2011). https://doi.org/10.1007/s11515-011-1118-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11515-011-1118-6