Abstract
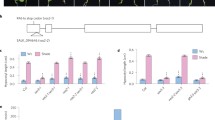
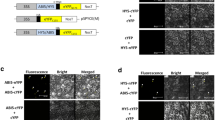
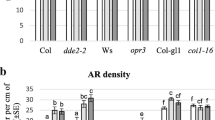
Light acts as the pivotal external environment cue to modulate plant growth and development. Seeds germinate in the soil without light to undergo skotomorphogenesis with rapidly elongating hypocotyls that facilitate emergence from the soil, while seedlings upon light exposure undergo photomorphogenesis with significantly inhibited hypocotyl elongation that benefits plants to stand up firmly and cope with the changing environment. In this study, we demonstrate that light promotes jasmonate (JA) biosynthesis to inhibit hypocotyl elongation and orchestrate seedling photomorphogenesis in Arabidopsis. We showed that JAinhibition on hypocotyl elongation is dependent on JA receptor COI1 and signaling components such as repressor proteins JAZs and transcription activators MYC2/MYC3/MYC4. Furthermore, we found that MYC2/MYC3/MYC4 activate the expression of photomorphogenesis regulator HY5 to repress cell elongation-related genes (such as SAUR62 and EXP2) essential for seedling photomorphogenesis. Our findings provide a novel insight into molecular mechanisms underlying how plants integrate light signal with hormone pathway to establish seedling photomorphogenesis.
Similar content being viewed by others
References
Acosta, I.F., Gasperini, D., Chételat, A., Stolz, S., Santuari, L., and Farmer, E.E. (2013). Role of NINJA in root jasmonate signaling. Proc Natl Acad Sci USA 110, 15473–15478.
Ahmad, M., Jarillo, J.A., Smirnova, O., and Cashmore, A.R. (1998). Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature 392, 720–723.
Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1, 213–222.
Boter, M., Ruíz-Rivero, O., Abdeen, A., and Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18, 1577–1591.
Browse, J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60, 183–205.
Chakraborty, M., Gangappa, S.N., Maurya, J.P., Sethi, V., Srivastava, A.K., Singh, A., Dutta, S., Ojha, M., Gupta, N., Sengupta, M., et al. (2019). Functional interrelation of MYC 2 and HY 5 plays an important role in Arabidopsis seedling development. Plant J 99, 1080–1097.
Chao, D.Y., and Lin, H.X. (2010). The tricks plants use to reach appropriate light. Sci China Life Sci 53, 916–926.
Chen, M., and Chory, J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21, 664–671.
Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Annu Rev Genet 38, 87–117.
Chen, Q., Sun, J., Zhai, Q., Zhou, W., Qi, L., Xu, L., Wang, B., Chen, R., Jiang, H., Qi, J., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23, 3335–3352.
Chory, J. (1992). A genetic model for light-regulated seedling development in Arabidopsis. Development 115, 337–354.
Cosgrove, D.J. (2000). Loosening of plant cell walls by expansins. Nature 407, 321–326.
Dai, L., Xu, L., Huang, D., Li, X., Luo, K., and Guan, C. (2002). ASK1 physically interacts with COI1 and is required for male fertility in Arabidopsis. Sci China Ser C-Life Sci 45, 631–636.
Glauser G., and Wolfender J.L. (2013). A non-targeted approach for extended liquid chromatography-mass spectrometry profiling of free and esterified jasmonates after wounding. Methods Mol Biol 1011, 123–134.
Hardtke, C.S. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19, 4997–5006.
Hu, P., Zhou, W., Cheng, Z., Fan, M., Wang, L., and Xie, D. (2013). JAV1 controls jasmonate-regulated plant defense. Mol Cell 50, 504–515.
Huang, H., Wang, C., Tian, H., Sun, Y., Xie, D., and Song, S. (2014). Amino acid substitutions of GLY98, LEU245 and GLU543 in COI1 distinctively affect jasmonate-regulated male fertility in Arabidopsis. Sci China Life Sci 57, 145–154.
Jiao, Y., Lau, O.S., and Deng, X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8, 217–230.
Kami, C., Lorrain, S., Hornitschek, P., and Fankhauser, C. (2010). Lightregulated plant growth and development. Curr Top Dev Biol 91, 29–66.
Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., Tongprasit, W., Zhao, H., Lee, I., and Deng, X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749.
Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., Huq, E., and Quail, P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18, 1815–1823.
Li, T., Jia, K.P., Lian, H.L., Yang, X., Li, L., and Yang, H.Q. (2014). Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem Biophys Res Commun 454, 78–83.
Liu, H., Liu, B., Zhao, C., Pepper, M., and Lin, C. (2011). The action mechanisms of plant cryptochromes. Trends Plant Sci 16, 684–691.
Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466.
Paik, I., and Huq, E. (2019). Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin Cell Dev Biol 92, 114–121.
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., Fan, M., Peng, W., Ren, C., and Xie, D. (2011). The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814.
Qi, T., Wang, J., Huang, H., Liu, B., Gao, H., Liu, Y., Song, S., and Xie, D. (2015). Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27, 1634–1649.
Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3, 85–93.
Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157.
Reymond, P., Bodenhausen, N., Van Poecke, R.M.P., Krishnamurthy, V., Dicke, M., and Farmer, E.E. (2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16, 3132–3147.
Rowe, H.C., Walley, J.W., Corwin, J., Chan, E.K.F., Dehesh, K., and Kliebenstein, D.J. (2010). Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog 6, e1000861.
Shan, X., Zhang, Y., Peng, W., Wang, Z., and Xie, D. (2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot 60, 3849–3860.
Song, S., Huang, H., Gao, H., Wang, J., Wu, D., Liu, X., Yang, S., Zhai, Q., Li, C., Qi, T., et al. (2014). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26, 263–279.
Song, S., Qi, T., Huang, H., and Xie, D. (2013). Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol Plant 6, 1065–1073.
Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89, 6837–6840.
Su, J., Liu, B., Liao, J., Yang, Z., Lin, C., and Oka, Y. (2017). Coordination of cryptochrome and phytochrome signals in the regulation of plant light responses. Agronomy 7, 25.
Sun, N., Wang, J., Gao, Z., Dong, J., He, H., Terzaghi, W., Wei, N., Deng, X.W., and Chen, H. (2016). Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113, 6071–6076.
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665.
von Arnim, A.G., and Deng, X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cellspecific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045.
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111, 1021–1058.
Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J. S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768.
Xie, D., Feys, B., James, S., Nieto-Rostro, M., and Turner, J. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094.
Xu, X., Paik, I., Zhu, L., and Huq, E. (2015). Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci 20, 641–650.
Yan, C., Fan, M., Yang, M., Zhao, J., Zhang, W., Su, Y., Xiao, L., Deng, H., and Xie, D. (2018). Injury activates Ca2+/calmodulin-dependent phosphorylation of JAV1-JAZ8-WRKY51 complex for jasmonate biosynthesis. Mol Cell 70, 136–149.e7.
Yan, J., Zhang, C., Gu, M., Bai, Z., Zhang, W., Qi, T., Cheng, Z., Peng, W., Luo, H., Nan, F., et al. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236.
Yang, Z., Liu, B., Su, J., Liao, J., Lin, C., and Oka, Y. (2017). Cryptochromes orchestrate transcription regulation of diverse blue light responses in plants. Photochem Photobiol 93, 112–127.
Yuan, Z., and Zhang, D. (2015). Roles of jasmonate signalling in plant inflorescence and flower development. Curr Opin Plant Biol 27, 44–51.
Zhai, Q., Zhang, X., Wu, F., Feng, H., Deng, L., Xu, L., Zhang, M., Wang, Q., and Li, C. (2015). Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27, 2814–2828.
Zhang, H., He, H., Wang, X., Wang, X., Yang, X., Li, L., and Deng, X.W. (2011). Genome-wide mapping of the HY5-mediated genenetworks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65, 346–358.
Zheng, Y., Cui, X., Su, L., Fang, S., Chu, J., Gong, Q., Yang, J., and Zhu, Z. (2017). Jasmonate inhibits COP1 activity to suppress hypocotyl elongation and promote cotyledon opening in etiolated Arabidopsis seedlings. Plant J 90, 1144–1155.
Acknowledgements
We thank Dr. Deng X.W. (Peking University) for providing hy5-205, pifq mutant seeds; Dr. Yang H.Q. (Fudan University) for providing cry1/2, phyB-9, phyA-211 mutant seeds; and Lu Meng, Dr. Ran Du for their technical support. This work was supported by the National Key R&D Program of China (2016YFA0500501) and National Natural Science Foundation of China (31630085).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Compliance and ethics The author(s) declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yi, R., Yan, J. & Xie, D. Light promotes jasmonate biosynthesis to regulate photomorphogenesis in Arabidopsis. Sci. China Life Sci. 63, 943–952 (2020). https://doi.org/10.1007/s11427-019-1584-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-019-1584-4