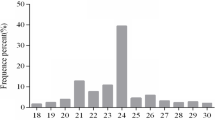
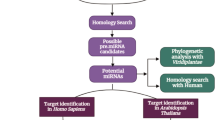
Abstract
Ophiocordyceps sinensis is well known as a traditional Chinese medicine and has widely been used for over 2,000 years to stimulate immune system, decrease blood pressure and to inhibit tumor growth. While miRNAs are increasingly recognized for their roles in post-transcriptional regulation of gene expression in animals and plants, miRNAs in fungi were less studied until the discovery of microRNA-like RNA (milRNA). High-throughput sequencing and bioinformatics approaches were used to identify conserved and novel milRNAs in O. sinensis. 40 conserved milRNAs were identified, while 23 pre-miRNA candidates encoding 31 novel milRNAs were predicted. Furthermore, the potential target genes of milRNAs in human were predicted and gene ontology analysis was applied to these genes. Enrichment analysis of GO-represented biological process showed that target genes of both conserved and novel milRNAs are involved in development, metabolic and immune processes, indicating the potential roles of milRNAs of O. sinensis in pharmacological effects as health food and traditional Chinese medicine. This study is the first report on genome-wide analysis of milRNAs in O. sinensis and it provides a useful resource to further study the potential roles of milRNAs as active components of O. sinensis in health food or traditional Chinese medicine.
Similar content being viewed by others
References
Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J. M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. (2000). Gene Ontology: tool for the unification of biology. Nat Genet 25, 25–29.
Bartel, D.P. (2004). microRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297.
Beatty, M., Guduric-Fuchs, J., Brown, E., Bridgett, S., Chakravarthy, U., Hogg, R.E., and Simpson, D.A. (2014). Small RNAs from plants, bacteria and fungi within the order Hypocreales are ubiquitous in human plasma. BMC Genomics 15, 933.
Bok, J.W., Lermer, L., Chilton, J., Klingeman, H.G., and Towers, G.H.N. (1999). Antitumor sterols from the mycelia of Cordyceps sinensis. Phytochemistry 51, 891–898.
Bonnet, E., Wuyts, J., Rouzé, P., and Van de Peer, Y. (2004). Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20, 2911–2917.
Borges, F., and Martienssen, R.A. (2015). The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16, 727–741.
Chen, R., Jiang, N., Jiang, Q., Sun, X., Wang, Y., Zhang, H., and Hu, Z. (2014). Exploring microRNA-like small RNAs in the filamentous fungus Fusarium oxysporum. PLoS ONE 9, e104956.
Chen, X., Gao, C., Li, H., Huang, L., Sun, Q., Dong, Y., Tian, C., Gao, S., Dong, H., Guan, D., et al. (2010). Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid and powdered milk products. Cell Res 20, 1128–1137.
Chin, A.R., Fong, M.Y., Somlo, G., Wu, J., Swiderski, P., Wu, X., and Wang, S.E. (2016). Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res 26, 217–228.
Dahlmann, T.A., and Kück, U. (2015). Dicer-dependent biogenesis of small RNAs and evidence for microRNA-like RNAs in the penicillin producing fungus Penicillium chrysogenum. PLoS ONE 10, e0125989.
Dumesic, P.A., Natarajan, P., Chen, C., Drinnenberg, I.A., Schiller, B.J., Thompson, J., Moresco, J.J., Yates Iii, J.R., Bartel, D.P., and Madhani, H.D. (2013). Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell 152, 957–968.
Enright, A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D.S. (2003). microRNA targets in Drosophila. Genome Biol 5, R1.
Friedländer, M.R., Chen, W., Adamidi, C., Maaskola, J., Einspanier, R., Knespel, S., and Rajewsky, N. (2008). Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26, 407–415.
Friedländer, M.R., Mackowiak, S.D., Li, N., Chen, W., and Rajewsky, N. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40, 37–52.
Griffiths-Jones, S. (2010). miRBase: microRNA sequences and annotation. Curr Protocols Bioinform 12, 1–10.
Huang, B.M., Ju, S.Y., Wu, C.S., Chuang, W.J., Sheu, C.C., and Leu, S.F. (2001). Cordyceps sinensis and its fractions stimulate MA-10 mouse Leydig tumor cell steroidogenesis. J Androl 22, 831–837.
Huang, B.M., Hsiao, K.Y., Chuang, P.C., Wu, M.H., Pan, H.A., and Tsai, S. J. (2004). Upregulation of steroidogenic enzymes and ovarian 17ß-estradiol in human granulosa-lutein cells by Cordyceps sinensis mycelium. Biol Reprod 70, 1358–1364.
Jiang, N., Yang, Y., Janbon, G., Pan, J., and Zhu, X. (2012). Identification and functional demonstration of miRNAs in the fungus Cryptococcus neoformans. PLoS ONE 7, e52734.
John, B., Enright, A.J., Aravin, A., Tuschl, T., Sander, C., and Marks, D.S. (2005). Correction: human microRNA targets. PLoS Biol 3, e264–1328.
Kang, K., Zhong, J., Jiang, L., Liu, G., Gou, C.Y., Wu, Q., Wang, Y., Luo, J., and Gou, D. (2013). Identification of microRNA-like RNAs in the filamentous fungus Trichoderma reesei by solexa sequencing. PLoS ONE 8, e76288.
Kuo, C.F., Chen, C.C., Luo, Y.H., Huang, R.Y., Chuang, W.J., Sheu, C.C., and Lin, Y.S. (2005). Cordyceps sinensis mycelium protects mice from group A streptococcal infection. J Med Microbiol 54, 795–802.
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25.
Lau, S.K.P., Chow, W.N., Wong, A.Y.P., Yeung, J.M.Y., Bao, J., Zhang, N., Lok, S., Woo, P.C.Y., and Yuen, K.Y. (2013). Identification of micro-RNA-like RNAs in mycelial and yeast phases of the thermal dimorphic fungus Penicillium marneffei. PLoS Negl Trop Dis 7, e2398.
Li, Y., Hsiang, T., Yang, R.H., Hu, X.D., Wang, K., Wang, W.J., Wang, X. L., Jiao, L., and Yao, Y.J. (2016). Comparison of different sequencing and assembly strategies for a repeat-rich fungal genome, Ophiocordyceps sinensis. J Microbiol Methods 128, 1–6.
Liang, G.F., Zhu, Y.L., Sun, B., Shao, Y.H., Jing, A.H., Wang, J.H., and Xiao, Z.D. (2014). Assessing the survival of exogenous plant micro-RNA in mice. Food Sci Nutr 2, 380–388.
Liu, C., Lu, S., and Ji, M.R. (1992). Effects of Cordyceps sinensis (CS) on in vitro natural killer cells (in Chinese). Zhongguo Zhong Xi Yi Jie He Za Zhi 12, 267–269, 259.
Lorenz, R., Bernhart, S.H., Höner Zu Siederdissen, C., Tafer, H., Flamm, C., Stadler, P.F., and Hofacker, I.L. (2011). ViennaRNA package 2.0. Algorithms Mol Biol 6, 26.
Manabe, N., Sugimoto, M., Azuma, Y., Taketomo, N., Yamashita, A., Tsuboi, H., Tsunoo, A., Kinjo, N., Nian-Lai, H., and Miyamoto, H. (1996). Effects of the mycelial extract of cultured Cordyceps sinensis on in vivo hepatic energy metabolism in the mouse. Jpn J Pharmacol 70, 85–88.
Manabe, N., Azuma, Y., Sugimoto, M., Uchio, K., Miyamoto, M., Taketomo, N., Tsuchita, H., and Miyamoto, H. (2000). Effects of the mycelial extract of cultured Cordyceps sinensis on in vivo hepatic energy metabolism and blood flow in dietary hypoferric anaemic mice. Br J Nutr 83, 197–204.
Mi, H., Huang, X., Muruganujan, A., Tang, H., Mills, C., Kang, D., and Thomas, P.D. (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183–D189.
Mlotshwa, S., Pruss, G.J., MacArthur, J.L., Endres, M.W., Davis, C., Hofseth, L.J., Peña, M.M., and Vance, V. (2015). A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res 25, 521–524.
Nakayashiki, H., and Nguyen, Q.B. (2008). RNA interference: roles in fungal biology. Curr Opin Microbiol 11, 494–502.
Nawrocki, E.P., Burge, S.W., Bateman, A., Daub, J., Eberhardt, R.Y., Eddy, S.R., Floden, E.W., Gardner, P.P., Jones, T.A., Tate, J., et al. (2015). Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43, D130–D137.
White, J.L., and Dawson, W.O. (1979). Effect of cordycepin triphosphate on in vitro RNA synthesis by plant viral replicases. J Virol 29, 811–814.
Winkler, D. (2008). Yartsa Gunbu (Cordyceps sinensis) and the fungal commodification of Tibet’s rural economy. Econ Bot 62, 291–305.
Wu, J., Wang, D., Liu, Y., Wang, L., Qiao, X., and Zhang, S. (2014). Identification of miRNAs involved in pear fruit development and quality. BMC Genomics 15, 953.
Xu, R.H., Peng, X.E., Chen, G.Z., and Chen, G.L. (1992). Effects of cordyceps sinensis on natural killer activity and colony formation of B16 melanoma. Chin Med J (Engl) 105, 97–101.
Yang, J., Farmer, L.M., Agyekum, A.A.A., Elbaz-Younes, I., and Hirschi, K.D. (2015a). Detection of an abundant plant-based small RNA in healthy consumers. PLoS ONE 10, e0137516.
Yang, J., Farmer, L.M., Agyekum, A.A.A., and Hirschi, K.D. (2015b). Detection of dietary plant-based small RNAs in animals. Cell Res 25, 517–520.
Yoshida, J., Takamura, S., Yamaguchi, N., Ren, L.J., Chen, H., Koshimura, S., and Suzuki, S. (1989). Antitumor activity of an extract of Cordyceps sinensis (Berk.) Sacc. against murine tumor cell lines. Jpn J Exp Med 59, 157–161.
Zhang, L., Hou, D., Chen, X., Li, D., Zhu, L., Zhang, Y., Li, J., Bian, Z., Liang, X., Cai, X., et al. (2012). Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 22, 273–274.
Zhou, J., Fu, Y., Xie, J., Li, B., Jiang, D., Li, G., and Cheng, J. (2012a). Identification of microRNA-like RNAs in a plant pathogenic fungus Sclerotinia sclerotiorum by high-throughput sequencing. Mol Genet Genomics 287, 275–282.
Zhou, Q., Wang, Z., Zhang, J., Meng, H., and Huang, B. (2012b). Genomewide identification and profiling of microRNA-like RNAs from Metarhizium anisopliae during development. Fungal Biol 116, 1156–1162.
Zhou, X.W., Li, L.J., and Tian, E.W. (2014). Advances in research of the artificial cultivation of Ophiocordyceps sinensis in China. Crit Rev Biotech 34, 233–243.
Zhou, Z., Li, X., Liu, J., Dong, L., Chen, Q., Liu, J., Kong, H., Zhang, Q., Qi, X., Hou, D., et al. (2015). Honeysuckle-encoded atypical micro-RNA2911 directly targets influenza A viruses. Cell Res 25, 39–49.
Zhu, J.S., Halpern, G.M., and Jones, K. (1998). The scientific rediscovery of a precious ancient Chinese herbal regimen: Cordyceps sinensis part II. J Altern Complement Med 4, 429–457.
Acknowledgements
This work was supported by the National Basic Research Program of China (2014CB542300), the National Natural Science Foundation of China (81602697), the Natural Science Foundation of Jiangsu Province (BE2016737) and the Fundamental Research Funds for the Central Universities (020814380070).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhang, W., Li, X., Ma, L. et al. Identification of microRNA-like RNAs in Ophiocordyceps sinensis. Sci. China Life Sci. 62, 349–356 (2019). https://doi.org/10.1007/s11427-017-9277-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-017-9277-9