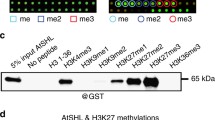
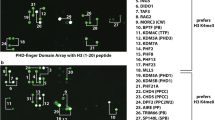
Abstract
In eukaryotes, epigenetic-based mechanisms are involved in almost all the important biological processes. Amongst different epigenetic regulation pathways, the dynamic covalent modifications on histones are the most extensively investigated and characterized types. The covalent modifications on histone can be “read” by specific protein domains and then subsequently trigger downstream signaling events. Plants generally possess epigenetic regulation systems similar to animals and fungi, but also exhibit some plant-specific features. Similar to animals and fungi, plants require distinct protein domains to specifically “read” modified histones in both modification-specific and sequence-specific manners. In this review, we will focus on recent progress of the structural studies on the recognition of the epigenetic marks on histones by plant reader proteins, and further summarize the general and exceptional features of plant histone mark readers.
Similar content being viewed by others
References
Aladjem, M.I. (2007). Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet 8, 588–600.
Andrews, F.H., Tong, Q., Sullivan, K.D., Cornett, E.M., Zhang, Y., Ali, M., Ahn, J.W., Pandey, A., Guo, A.H., Strahl, B.D., Costello, J.C., Espinosa, J.M., Rothbart, S.B., and Kutateladze, T.G. (2016). Multivalent chromatin engagement and inter-domain crosstalk regulate MORC3 ATPase. Cell Rep 16, 3195–3207.
Bergamin, E., Sarvan, S., Malette, J., Eram, M.S., Yeung, S., Mongeon, V., Joshi, M., Brunzelle, J.S., Michaels, S.D., Blais, A., Vedadi, M., and Couture, J.F. (2017). Molecular basis for the methylation specificity of ATXR5 for histone H3. Nucleic Acids Res 45, 6375–6387.
Berger, S.L., Kouzarides, T., Shiekhattar, R., and Shilatifard, A. (2009). An operational definition of epigenetics. Genes Dev 23, 781–783.
Blus, B.J., Wiggins, K., and Khorasanizadeh, S. (2011). Epigenetic virtues of chromodomains. Crit Rev Biochem Mol Biol 46, 507–526.
Bu, Z., Yu, Y., Li, Z., Liu, Y., Jiang, W., Huang, Y., and Dong, A.W. (2014). Regulation of Arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet 10, e1004617.
Costas, C., de la Paz Sanchez, M., Stroud, H., Yu, Y., Oliveros, J.C., Feng, S., Benguria, A., López-Vidriero, I., Zhang, X., Solano, R., Jacobsen, S.E., and Gutierrez, C. (2011). Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat Struct Mol Biol 18, 395–400.
de la Paz Sanchez, M., and Gutierrez, C. (2009). Arabidopsis ORC1 is a PHD-containing H3K4me3 effector that regulates transcription. Proc Natl Acad Sci USA 106, 2065–2070.
Du, J. (2016). Structure and mechanism of plant DNA methyltransferases. Adv Exp Med Biol 945, 173–192.
Du, J., Johnson, L.M., Jacobsen, S.E., and Patel, D.J. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol 16, 519–532.
Du, J., Zhong, X., Bernatavichute, Y.V., Stroud, H., Feng, S., Caro, E., Vashisht, A.A., Terragni, J., Chin, H.G., Tu, A., Hetzel, J., Wohlschlegel, J.A., Pradhan, S., Patel, D.J., and Jacobsen, S.E. (2012). Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151, 167–180.
Duncker, B.P., Chesnokov, I.N., and McConkey, B.J. (2009). The origin recognition complex protein family. Genome Biol 10, 214.
Hoppmann, V., Thorstensen, T., Kristiansen, P.E., Veiseth, S.V., Rahman, M.A., Finne, K., Aalen, R.B., and Aasland, R. (2011). The CW domain, a new histone recognition module in chromatin proteins. EMBO J 30, 1939–1952.
Hou, Z., Bernstein, D.A., Fox, C.A., and Keck, J.L. (2005). Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc Natl Acad Sci USA 102, 8489–8494.
Hsu, H.C., Stillman, B., and Xu, R.M. (2005). Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc Natl Acad Sci USA 102, 8519–8524.
Jacob, Y., Bergamin, E., Donoghue, M.T.A., Mongeon, V., LeBlanc, C., Voigt, P., Underwood, C.J., Brunzelle, J.S., Michaels, S.D., Reinberg, D., Couture, J.F., and Martienssen, R.A. (2014). Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343, 1249–1253.
Jacob, Y., Feng, S., LeBlanc, C.A., Bernatavichute, Y.V., Stroud, H., Cokus, S., Johnson, L.M., Pellegrini, M., Jacobsen, S.E., and Michaels, S.D. (2009). ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16, 763–768.
Jacob, Y., Stroud, H., Leblanc, C., Feng, S., Zhuo, L., Caro, E., Hassel, C., Gutierrez, C., Michaels, S.D., and Jacobsen, S.E. (2010). Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466, 987–991.
Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293, 1074–1080.
Johnson, L.M., Du, J., Hale, C.J., Bischof, S., Feng, S., Chodavarapu, R.K., Zhong, X., Marson, G., Pellegrini, M., Segal, D.J., Patel, D.J., and Jacobsen, S.E. (2014). SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507, 124–128.
Kolasinska-Zwierz, P., Down, T., Latorre, I., Liu, T., Liu, X.S., and Ahringer, J. (2009). Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41, 376–381.
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705.
Kuo, A.J., Song, J., Cheung, P., Ishibe-Murakami, S., Yamazoe, S., Chen, J.K., Patel, D.J., and Gozani, O. (2012). The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484, 115–119.
Larschan, E., Alekseyenko, A.A., Gortchakov, A.A., Peng, S., Li, B., Yang, P., Workman, J.L., Park, P.J., and Kuroda, M.I. (2007). MSL complex is attracted to genes marked by H3K36 trimethylation using a sequenceindependent mechanism. Mol Cell 28, 121–133.
Law, J.A., Du, J., Hale, C.J., Feng, S., Krajewski, K., Palanca, A.M.S., Strahl, B.D., Patel, D.J., and Jacobsen, S.E. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389.
Law, J.A., Vashisht, A.A., Wohlschlegel, J.A., and Jacobsen, S.E. (2011). SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet 7, e1002195.
Li, M., and Fang, Y.D. (2015). Histone variants: the artists of eukaryotic chromatin. Sci China Life Sci 58, 232–239.
Li, S., Yang, Z., Du, X., Liu, R., Wilkinson, A.W., Gozani, O., Jacobsen, S.E., Patel, D.J., and Du, J. (2016a). Structural basis for the unique multivalent readout of unmodified H3 tail by Arabidopsis ORC1b BAH-PHD cassette. Structure 24, 486–494.
Li, S., Yen, L., Pastor, W.A., Johnston, J.B., Du, J., Shew, C.J., Liu, W., Ho, J., Stender, B., Clark, A.T., Burlingame, A.L., Daxinger, L., Patel, D.J., and Jacobsen, S.E. (2016b). Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin. Proc Natl Acad Sci USA 113, E5108–E5116.
Li, Y., and Li, H. (2012). Many keys to push: diversifying the ‘readership’ of plant homeodomain fingers. Acta Biochim Biophys Sin 44, 28–39.
Li, Y., Zhao, D., Chen, Z., and Li, H. (2017). YEATS domain: Linking histone crotonylation to gene regulation. Transcription 8, 9–14.
Liu, J., Bai, G., Zhang, C., Chen, W., Zhou, J., Zhang, S., Chen, Q., Deng, X., He, X.J., and Zhu, J.K. (2011). An atypical component of RNA-directed DNA methylation machinery has both DNA methylation-dependent and-independent roles in locus-specific transcriptional gene silencing. Cell Res 21, 1691–1700.
Liu, Y., and Min, J. (2016). Structure and function of histone methylationbinding proteins in plants. Biochem J 473, 1663–1680.
Liu, Y., Tempel, W., Zhang, Q., Liang, X., Loppnau, P., Qin, S., and Min, J. (2016a). Family-wide characterization of histone binding abilities of human CW domain-containing proteins. J Biol Chem 291, 9000–9013.
Liu, Y., Wu, H., Yu, Y., and Huang, Y. (2016b). Structural studies on MRG701 chromodomain reveal a novel dimerization interface of MRG proteins in green plants. Protein Cell 7, 792–803.
Matzke, M.A., Kanno, T., and Matzke, A.J.M. (2015). RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66, 243–267.
Matzke, M.A., and Mosher, R.A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15, 394–408.
Mukherjee, K., Brocchieri, L., and Bürglin, T.R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol 26, 2775–2794.
Nady, N., Lemak, A., Walker, J.R., Avvakumov, G.V., Kareta, M.S., Achour, M., Xue, S., Duan, S., Allali-Hassani, A., Zuo, X., Wang, Y.X., Bronner, C., Chédin, F., Arrowsmith, C.H., and Dhe-Paganon, S. (2011). Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem 286, 24300–24311.
Patel, D.J. (2016). A structural perspective on readout of epigenetic histone and DNA methylation marks. Cold Spring Harb Perspect Biol 8, a018754.
Patel, D.J., and Wang, Z. (2013). Readout of epigenetic modifications. Annu Rev Biochem 82, 81–118.
Perry, J., and Zhao, Y. (2003). The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem Sci 28, 576–580.
Roudier, F., Ahmed, I., Bérard, C., Sarazin, A., Mary-Huard, T., Cortijo, S., Bouyer, D., Caillieux, E., Duvernois-Berthet, E., Al-Shikhley, L., Giraut, L., Després, B., Drevensek, S., Barneche, F., Dèrozier, S., Brunaud, V., Aubourg, S., Schnittger, A., Bowler, C., Martin-Magniette, M.L., Robin, S., Caboche, M., and Colot, V. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30, 1928–1938.
Stroud, H., Do, T., Du, J., Zhong, X., Feng, S., Johnson, L., Patel, D.J., and Jacobsen, S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21, 64–72.
Sun, B., Hong, J., Zhang, P., Dong, X., Shen, X., Lin, D., and Ding, J. (2008). Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J Biol Chem 283, 36504–36512.
Xu, C., Cui, G., Botuyan, M.V., and Mer, G. (2008). Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure 16, 1740–1750.
Yang, N., and Xu, R.M. (2013). Structure and function of the BAH domain in chromatin biology. Crit Rev Biochem Mol Biol 48, 211–221.
Zhang, H., Ma, Z.Y., Zeng, L., Tanaka, K., Zhang, C.J., Ma, J., Bai, G., Wang, P., Zhang, S.W., Liu, Z.W., Cai, T., Tang, K., Liu, R., Shi, X., He, X.J., and Zhu, J.K. (2013). DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci USA 110, 8290–8295.
Zhang, P., Du, J., Sun, B., Dong, X., Xu, G., Zhou, J., Huang, Q., Liu, Q., Hao, Q., and Ding, J. (2006). Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res 34, 6621–6628.
Acknowledgements
This work was supported by the National Key R&D Program of China (2016YFA0503200), National Natural Science Foundation of China (31622032), and the Chinese Academy of Sciences to Jiamu Du.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, R., Li, X., Chen, W. et al. Structure and mechanism of plant histone mark readers. Sci. China Life Sci. 61, 170–177 (2018). https://doi.org/10.1007/s11427-017-9163-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-017-9163-4