Abstract
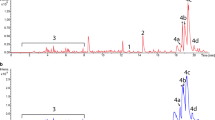
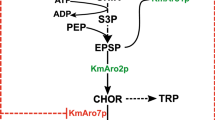
An ideal surrogate host for heterologous production of various natural products is expected to have efficient nutrient utilization, fast growth, abundant precursors and energy supply, and a pronounced gene expression. Streptomyces albus BK3-25 is a high-yield industrial strain producing type-I polyketide salinomycin, with a unique ability of bean oil utilization. Its potential of being a surrogate host for heterologous production of PKS was engineered and evaluated herein. Firstly, introduction of a three-gene cassette for the biosynthesis of ethylmalonyl-CoA resulted in accumulation of ethylmalonyl-CoA precursor and salinomycin, and subsequent deletion of the salinomycin biosynthetic gene cluster resulted in a host with rich supplies of common polyketide precursors, including malonyl-CoA, methylmalonyl-CoA, and ethylmalonyl-CoA. Secondly, the energy and reducing force were measured, and the improved accumulation of ATP and NADPH was observed in the mutant. Furthermore, the strength of a series of selected endogenous promoters based on microarray data was assessed at different growth phases, and a strong constitutive promoter was identified, providing a useful tool for further engineered gene expression. Finally, the potential of the BK3-25 derived host ZXJ-6 was evaluated with the introduction of the actinorhodin biosynthetic gene cluster from Streptomyces coelicolor, and the heterologous production of actinorhodin was obtained. This work clearly indicated the potential of the high-yield salinomycin producer as a surrogate host for heterologous production of polyketides, although more genetic manipulation should be conducted to streamline its performance.
Similar content being viewed by others
References
Armando, J.W., Boghigian, B.A., and Pfeifer, B.A. (2012). LC-MS/MS quantification of short-chain acyl-CoA’s in Escherichia coli demonstrates versatile propionyl-CoA synthetase substrate specificity. Lett Appl Microbiol 54, 140–148.
Barkei, J.J., Kevany, B.M., Felnagle, E.A., and Thomas, M.G. (2009). Investigations into viomycin biosynthesis by using heterologous production in Streptomyces lividans. ChemBioChem 10, 366–376.
Beites, T., and Mendes, M.V. (2015). Chassis optimization as a cornerstone for the application of synthetic biology based strategies in microbial secondary metabolism. Front Microbiol 6, 906.
Bérdy, J. (2005). Bioactive microbial metabolites. J Antibiot 58, 1–26.
Eustáquio, A.S., Gust, B., Li, S.M., Pelzer, S., Wohlleben, W., Chater, K.F., and Heide, L. (2004). Production of 8′-halogenated and 8′-unsubstituted novobiocin derivatives in genetically engineered Streptomyces coelicolor strains. Chem Biol 11, 1561–1572.
Felnagle, E.A., Rondon, M.R., Berti, A.D., Crosby, H.A., and Thomas, M.G. (2007). Identification of the biosynthetic gene cluster and an additional gene for resistance to the antituberculosis drug capreomycin. Appl Environ Microbiol 73, 4162–4170.
Gullón, S., Olano, C., Abdelfattah, M.S., Braña, A.F., Rohr, J., Méndez, C., and Salas, J.A. (2006). Isolation, characterization, and heterologous expression of the biosynthesis gene cluster for the antitumor anthracycline steffimycin. Appl Environ Microbiol 72, 4172–4183.
Hammer, K., Mijakovic, I., and Jensen, P.R. (2006). Synthetic promoter libraries—tuning of gene expression. Trends Biotech 24, 53–55.
Hara, M., Asano, K., Kawamoto, I., Takiouchi, T., Katsumata, S., Takahashi, K.I., and Nakano, H. (1989). Leinamycin, a new antitumor antibiotic from Streptomyces: producing organism, fermentation and isolation. J Antibiot 42, 1768–1774.
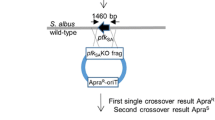
He, Y. (2010). Two pHZ1358 derivative vectors for efficient gene knockout in Streptomyces. J Microbiol Biotechnol 20, 678–682.
Huang, J., Yu, Z., Li, M.H., Wang, J.D., Bai, H., Zhou, J., and Zheng, Y.G. (2016). High level of spinosad production in the heterologous host Saccharopolyspora erythraea. Appl Environ Microbiol 82, 5603–5611.
Jiang, C., Wang, H., Kang, Q., Liu, J., and Bai, L. (2011). Cloning and characterization of the polyether salinomycin biosynthesis gene cluster of Streptomyces albus XM211. Appl Environ Microbiol 78, 994–1003.
Jung, W.S., Kim, E., Yoo, Y.J., Ban, Y.H., Kim, E.J., and Yoon, Y.J. (2014). Characterization and engineering of the ethylmalonyl-CoA pathway towards the improved heterologous production of polyketides in Streptomyces venezuelae. Appl Microbiol Biotechnol 98, 3701–3713.
Kao, C.M., Katz, L., and Khosla, C. (1994). Engineered biosynthesis of a complete macrolactone in a heterologous host. Science 265, 509–512.
Komatsu, M., Komatsu, K., Koiwai, H., Yamada, Y., Kozone, I., Izumikawa, M., Hashimoto, J., Takagi, M., Omura, S., Shin-ya, K., Cane, D.E., and Ikeda, H. (2013). EngineeredStreptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol 2, 384–396.
Komatsu, M., Uchiyama, T., Omura, S., Cane, D.E., and Ikeda, H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA 107, 2646–2651.
Liu, Q., Xiao, L., Zhou, Y., Deng, K., Tan, G., Han, Y., Liu, X., Deng, Z., and Liu, T. (2016). Development of Streptomyces sp. FR-008 as an emerging chassis. Synth Syst Biotechnol 1, 1–8.
Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408.
Lu, C., Wu, H., Su, X., and Bai, L. (2017). Elimination of indigenous linear plasmids in Streptomyces hygroscopicus var. jinggangensis and Streptomyces sp. FR008 to increase validamycin A and candicidin productivities. Appl Microbiol Biotechnol 101, 4247–4257.
Lu, C., Zhang, X., Jiang, M., and Bai, L. (2016). Enhanced salinomycin production by adjusting the supply of polyketide extender units in Streptomyces albus. Metab Eng 35, 129–137.
Luo, Y., Li, B.Z., Liu, D., Zhang, L., Chen, Y., Jia, B., Zeng, B.X., Zhao, H., and Yuan, Y.J. (2015a). Engineered biosynthesis of natural products in heterologous hosts. Chem Soc Rev 44, 5265–5290.
Luo, Y., Zhang, L., Barton, K.W., and Zhao, H. (2015b). Systematic identification of a panel of strong constitutive promoters from Streptomyces albus. Acs Synth Biol 4, 209–223.
Miao, V., Coëffet-Legal, M.F., Brian, P., Brost, R., Penn, J., Whiting, A., Martin, S., Ford, R., Parr, I., Bouchard, M., Silva, C.J., Wrigley, S.K., and Baltz, R.H. (2005). Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151, 1507–1523.
Ongley, S.E., Bian, X., Neilan, B.A., and Müller, R. (2013). Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat Prod Rep 30, 1121–1138.
Paget M.S., Chamberlin L., Atrih A., Foster S.J., and Buttner M.J. (1999). Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J Bacteriol 181, 204–211.
Park, S.H., Kim, H.U., Kim, T.Y., Park, J.S., Kim, S.S., and Lee, S.Y. (2014). Metabolic engineering of Corynebacterium glutamicum for L-arginine production. Nat Commun 5, 4618.
Rafat, A., Mirita, F.W., Yvonne, T., Martin, H., Mohamed, M., Yousra, A., Arne, M., Sergii, K., Marco, J., and Markus, H. (2016). Post-translational serine/threonine phosphorylation and lysine acetylation: a novel regulatory aspect of the global nitrogen response regulator GlnR in S. coelicolor M145. Front Mol Biosci 3, 38.
Rodriguez, E., Banchio, C., Diacovich, L., Bibb, M.J., and Gramajo, H. (2001). Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor A3(2). Appl Environ Microbiol 67, 4166–4176.
Ryu, Y.G., Butler, M.J., Chater, K.F., and Lee, K.J. (2006). Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl Environ Microbiol 72, 7132–7139.
Sawers, R.G., Falke, D., and Fischer, M. (2016). Oxygen and nitrate respiration in Streptomyces coelicolor A3(2). Adv Microb Physiol 68, 1–40.
Shinkawa, H., Hatada, Y., Okada, M., Kinashi, H., and Nimi, O. (1995). Nucleotide sequence of a principal sigma factor gene (hrdB) of Streptomyces griseus. J Biochem 118, 494–499.
Siebenberg, S., Bapat, P.M., Lantz, A.E., Gust, B., and Heide, L. (2010). Reducing the variability of antibiotic production in Streptomyces by cultivation in 24-square deepwell plates. J Biosci Bioeng 109, 230–234.
Siegl, T., Tokovenko, B., Myronovskyi, M., and Luzhetskyy, A. (2013). Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab Eng 19, 98–106.
Wendt-Pienkowski, E., Huang, Y., Zhang, J., Li, B., Jiang, H., Kwon, H., Hutchinson, C.R., and Shen, B. (2005). Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster fromStreptomyces griseus. J Am Chem Soc 127, 16442–16452.
Zhang, J., Martin, C., Shifflet, M.A., Salmon, P., Brix, T., Greasham, R., Buckland, B., and Chartrain, M. (1996). Development of a defined medium fermentation process for physostigmine production by Streptomyces griseofuscus. Appl Microbiol Biotechnol 44, 568–575.
Zhang, X., Lu, C., and Bai, L. (2017). Mechanism of salinomycin overproduction in Streptomyces albus as revealed by comparative functional genomics. Appl Microbiol Biotechnol 101, 4635–4644.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (21661140002 and 31470157) and the Ministry of Science and Technology of China (2012CB721005 and 2012AA022107). We are grateful to Prof. Yongquan Li (Zhejiang University) and Zhejiang Shenghua Biok Biology Co., Ltd. for providing the strain S. albus BK3-25. We are grateful to Dr. Meifeng Tao (Shanghai Jiao Tong University) for providing the plasmid pMM1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, X., Lu, C. & Bai, L. Conversion of the high-yield salinomycin producer Streptomyces albus BK3-25 into a surrogate host for polyketide production. Sci. China Life Sci. 60, 1000–1009 (2017). https://doi.org/10.1007/s11427-017-9122-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-017-9122-8