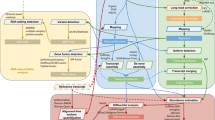
Abstract
Bioinformatics methods for various RNA-seq data analyses are in fast evolution with the improvement of sequencing technologies. However, many challenges still exist in how to efficiently process the RNA-seq data to obtain accurate and comprehensive results. Here we reviewed the strategies for improving diverse transcriptomic studies and the annotation of genetic variants based on RNA-seq data. Mapping RNA-seq reads to the genome and transcriptome represent two distinct methods for quantifying the expression of genes/transcripts. Besides the known genes annotated in current databases, many novel genes/transcripts (especially those long noncoding RNAs) still can be identified on the reference genome using RNA-seq. Moreover, owing to the incompleteness of current reference genomes, some novel genes are missing from them. Genome- guided and de novo transcriptome reconstruction are two effective and complementary strategies for identifying those novel genes/transcripts on or beyond the reference genome. In addition, integrating the genes of distinct databases to conduct transcriptomics and genetics studies can improve the results of corresponding analyses.
Article PDF
Similar content being viewed by others
References
Barrett, T., Wilhite, S.E., Ledoux, P., Evangelista, C., Kim, I.F., Tomashevsky, M., Marshall, K.A., Phillippy, K.H., Sherman, P.M., Holko, M., Yefanov, A., Lee, H., Zhang, N., Robertson, C.L., Serova, N., Davis, S., and Soboleva, A. (2013). NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41, D991–D995.
Cabili, M.N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., and Rinn, J.L. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927.
Chang, Z., Li, G., Liu, J., Zhang, Y., Ashby, C., Liu, D., Cramer, C.L., and Huang, X. (2015). Bridger: a new framework for de novo transcriptome assembly using RNA-seq data. Genome Biol 16, 30.
Chen, G., Li, R., Shi, L., Qi, J., Hu, P., Luo, J., Liu, M., and Shi, T. (2011a). Revealing the missing expressed genes beyond the human reference genome by RNA-Seq. BMC Genomics 12, 590.
Chen, G., Wang, C., Shi, L., Qu, X., Chen, J., Yang, J., Shi, C., Chen, L., Zhou, P., Ning, B., Tong, W., and Shi, T. (2013a). Incorporating the human gene annotations in different databases significantly improved transcriptomic and genetic analyses. RNA 19, 479–489.
Chen, G., Wang, C., Shi, L., Tong, W., Qu, X., Chen, J., Yang, J., Shi, C., Chen, L., Zhou, P., Lu, B., and Shi, T. (2013b). Comprehensively identifying and characterizing the missing gene sequences in human reference genome with integrated analytic approaches. Hum Genet 132, 899–911.
Chen, G., Wang, C., and Shi, T. (2011b). Overview of available methods for diverse RNA-Seq data analyses. Sci China Life Sci 54, 1121–1128.
Chen, G., Yin, K., Shi, L., Fang, Y., Qi, Y., Li, P., Luo, J., He, B., Liu, M., and Shi, T. (2011c). Comparative analysis of human protein-coding and noncoding RNAs between brain and 10 mixed cell lines by RNA-Seq. PLoS One 6, e28318.
Chen, G., Yu, D., Chen, J., Cao, R., Yang, J., Wang, H., Ji, X., Ning, B., and Shi, T. (2015). Re-annotation of presumed noncoding disease/ trait-associated genetic variants by integrative analyses. Sci Rep 5, 9453.
Chettoor, A.M., Givan, S.A., Cole, R.A., Coker, C.T., Unger-Wallace, E., Vejlupkova, Z., Vollbrecht, E., Fowler, J.E., and Evans, M.M. (2014). Discovery of novel transcripts and gametophytic functions via RNA-seq analysis of maize gametophytic transcriptomes. Genome Biol 15, 414.
Consortium, E.P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74.
Cunningham, F., Amode, M.R., Barrell, D., Beal, K., Billis, K., Brent, S., Carvalho-Silva, D., Clapham, P., Coates, G., Fitzgerald, S., Gil, L., Giron, C.G., Gordon, L., Hourlier, T., Hunt, S.E., Janacek, S.H., Johnson, N., Juettemann, T., Kahari, A.K., Keenan, S., Martin, F.J., Maurel, T., McLaren, W., Murphy, D.N., Nag, R., Overduin, B., Parker, A., Patricio, M., Perry, E., Pignatelli, M., Riat, H.S., Sheppard, D., Taylor, K., Thormann, A., Vullo, A., Wilder, S.P., Zadissa, A., Aken, B.L., Birney, E., Harrow, J., Kinsella, R., Muffato, M., Ruffier, M., Searle, S.M., Spudich, G., Trevanion, S.J., Yates, A., Zerbino, D.R., and Flicek, P. (2015). Ensembl 2015. Nucleic Acids Res 43, D662–669.
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., Guernec, G., Martin, D., Merkel, A., Knowles, D.G., Lagarde, J., Veeravalli, L., Ruan, X., Ruan, Y., Lassmann, T., Carninci, P., Brown, J.B., Lipovich, L., Gonzalez, J.M., Thomas, M., Davis, C.A., Shiekhattar, R., Gingeras, T.R., Hubbard, T.J., Notredame, C., Harrow, J., and Guigo, R. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789.
Dobin, A., Davis, C.A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M., and Gingeras, T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21.
Engstrom, P.G., Steijger, T., Sipos, B., Grant, G.R., Kahles, A., Ratsch, G., Goldman, N., Hubbard, T.J., Harrow, J., Guigo, R., Bertone, P., and Consortium, R. (2013). Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods 10, 1185–1191.
Fan, X.N., and Zhang, S.W. (2015). lncRNA-MFDL: identification of human long non-coding RNAs by fusing multiple features and using deep learning. Mol Biosyst 11, 892–897.
Fonseca, N.A., Rung, J., Brazma, A., and Marioni, J.C. (2012). Tools for mapping high-throughput sequencing data. Bioinformatics 28, 3169–3177.
Garber, M., Grabherr, M.G., Guttman, M., and Trapnell, C. (2011). Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods 8, 469–477.
Gongora-Castillo, E., and Buell, C.R. (2013). Bioinformatics challenges in de novo transcriptome assembly using short read sequences in the absence of a reference genome sequence. Nat Prod Rep 30, 490–500.
Grabherr, M.G., Haas, B.J., Yassour, M., Levin, J.Z., Thompson, D.A., Amit, I., Adiconis, X., Fan, L., Raychowdhury, R., Zeng, Q., Chen, Z., Mauceli, E., Hacohen, N., Gnirke, A., Rhind, N., di Palma, F., Birren, B.W., Nusbaum, C., Lindblad-Toh, K., Friedman, N., and Regev, A. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29, 644–652.
Guttman, M., Garber, M., Levin, J.Z., Donaghey, J., Robinson, J., Adiconis, X., Fan, L., Koziol, M.J., Gnirke, A., Nusbaum, C., Rinn, J.L., Lander, E.S., and Regev, A. (2010). Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi- exonic structure of lincRNAs. Nature Biotechnol 28, 503–510.
Harrow, J., Frankish, A., Gonzalez, J.M., Tapanari, E., Diekhans, M., Kokocinski, F., Aken, B.L., Barrell, D., Zadissa, A., Searle, S., Barnes, I., Bignell, A., Boychenko, V., Hunt, T., Kay, M., Mukherjee, G., Rajan, J., Despacio-Reyes, G., Saunders, G., Steward, C., Harte, R., Lin, M., Howald, C., Tanzer, A., Derrien, T., Chrast, J., Walters, N., Balasubramanian, S., Pei, B., Tress, M., Rodriguez, J.M., Ezkurdia, I., van Baren, J., Brent, M., Haussler, D., Kellis, M., Valencia, A., Reymond, A., Gerstein, M., Guigo, R., and Hubbard, T.J. (2012). GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22, 1760–1774.
Jiang, H., and Wong, W.H. (2009). Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25, 1026–1032.
Kent, W.J. (2002). BLAT—the BLAST-like alignment tool. Genome Res 12, 656–664.
Kielbasa, S.M., Wan, R., Sato, K., Horton, P., and Frith, M.C. (2011). Adaptive seeds tame genomic sequence comparison. Genome Res 21, 487–493.
Kim, D., Langmead, B., and Salzberg, S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360.
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36.
Kodama, Y., Shumway, M., Leinonen, R., and International Nucleotide Sequence Database, C. (2012). The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res 40, D54–D56.
Kolesnikov, N., Hastings, E., Keays, M., Melnichuk, O., Tang, Y.A., Williams, E., Dylag, M., Kurbatova, N., Brandizi, M., Burdett, T., Megy, K., Pilicheva, E., Rustici, G., Tikhonov, A., Parkinson, H., Petryszak, R., Sarkans, U., and Brazma, A. (2015). ArrayExpress update— simplifying data submissions. Nucleic Acids Res 43, D1113–D1116.
Kong, L., Zhang, Y., Ye, Z.Q., Liu, X.Q., Zhao, S.Q., Wei, L., and Gao, G. (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res, W345–W349.
Konig, J., Zarnack, K., Luscombe, N.M., and Ule, J. (2011). Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet 13, 77–83.
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559.
Lee, C., and Kikyo, N. (2012). Strategies to identify long noncoding RNAs involved in gene regulation. Cell Biosci 2, 37.
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760.
Li, H., and Homer, N. (2010). A survey of sequence alignment algorithms for next-generation sequencing. Briefings Bioinform 11, 473–483.
Li, J.H., Liu, S., Zhou, H., Qu, L.H., and Yang, J.H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42, D92–97.
Li, R., Li, Y., Zheng, H., Luo, R., Zhu, H., Li, Q., Qian, W., Ren, Y., Tian, G., Li, J., Zhou, G., Zhu, X., Wu, H., Qin, J., Jin, X., Li, D., Cao, H., Hu, X., Blanche, H., Cann, H., Zhang, X., Li, S., Bolund, L., Kristiansen, K., Yang, H., Wang, J., and Wang, J. (2010). Building the sequence map of the human pan-genome. Nat Biotechnol 28, 57–63.
Liao, Q., Liu, C., Yuan, X., Kang, S., Miao, R., Xiao, H., Zhao, G., Luo, H., Bu, D., Zhao, H., Skogerbo, G., Wu, Z., and Zhao, Y. (2011). Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res 39, 3864–3878.
Liu, J., Gough, J., and Rost, B. (2006). Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genet 2, e29.
Martin, J.A., and Wang, Z. (2011). Next-generation transcriptome assembly. Nat Rev Genet 12, 671–682.
Nesvizhskii, A.I. (2007). Protein identification by tandem mass spectrometry and sequence database searching. Methods Mol Biol 367, 87–119.
Nielsen, R., Paul, J.S., Albrechtsen, A., and Song, Y.S. (2011). Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 12, 443–451.
Oshlack, A., Robinson, M.D., and Young, M.D. (2010). From RNA-seq reads to differential expression results. Genome Biol 11, 220.
Ozsolak, F., and Milos, P.M. (2011). RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12, 87–98.
Pauli, A., Valen, E., Lin, M.F., Garber, M., Vastenhouw, N.L., Levin, J.Z., Fan, L., Sandelin, A., Rinn, J.L., Regev, A., and Schier, A.F. (2012). Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22, 577–591.
Pepke, S., Wold, B., and Mortazavi, A. (2009). Computation for ChIP-seq and RNA-seq studies. Nat Methods 6, S22–32.
Pruitt, K.D., Brown, G.R., Hiatt, S.M., Thibaud-Nissen, F., Astashyn, A., Ermolaeva, O., Farrell, C.M., Hart, J., Landrum, M.J., McGarvey, K.M., Murphy, M.R., O’ Leary, N.A., Pujar, S., Rajput, B., Rangwala, S.H., Riddick, L.D., Shkeda, A., Sun, H., Tamez, P., Tully, R.E., Wallin, C., Webb, D., Weber, J., Wu, W., DiCuccio, M., Kitts, P., Maglott, D.R., Murphy, T.D., and Ostell, J.M. (2014). RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42, D756–D763.
Quek, X.C., Thomson, D.W., Maag, J.L., Bartonicek, N., Signal, B., Clark, M.B., Gloss, B.S., and Dinger, M.E. (2015). lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res 43, D168–D173.
Roberts, A., Pimentel, H., Trapnell, C., and Pachter, L. (2011). Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329.
Robertson, G., Schein, J., Chiu, R., Corbett, R., Field, M., Jackman, S.D., Mungall, K., Lee, S., Okada, H.M., Qian, J.Q., Griffith, M., Raymond, A., Thiessen, N., Cezard, T., Butterfield, Y.S., Newsome, R., Chan, S.K., She, R., Varhol, R., Kamoh, B., Prabhu, A.L., Tam, A., Zhao, Y., Moore, R.A., Hirst, M., Marra, M.A., Jones, S.J., Hoodless, P.A., and Birol, I. (2010). De novo assembly and analysis of RNA-seq data. Nat Methods 7, 909–912.
Rosenbloom, K.R., Armstrong, J., Barber, G.P., Casper, J., Clawson, H., Diekhans, M., Dreszer, T.R., Fujita, P.A., Guruvadoo, L., Haeussler, M., Harte, R.A., Heitner, S., Hickey, G., Hinrichs, A.S., Hubley, R., Karolchik, D., Learned, K., Lee, B.T., Li, C.H., Miga, K.H., Nguyen, N., Paten, B., Raney, B.J., Smit, A.F., Speir, M.L., Zweig, A.S., Haussler, D., Kuhn, R.M., and Kent, W.J. (2015). The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 43, D670–681.
Ruiz-Orera, J., Messeguer, X., Subirana, J.A., and Alba, M.M. (2014). Long non-coding RNAs as a source of new peptides. eLife 3, e03523.
Schulz, M.H., Zerbino, D.R., Vingron, M., and Birney, E. (2012). Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28, 1086–1092.
Thierry-Mieg, D., and Thierry-Mieg, J. (2006). AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol 7 Suppl 1, S12 11–14.
Trapnell, C., Williams, B.A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M.J., Salzberg, S.L., Wold, B.J., and Pachter, L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515.
Turro, E., Su, S.Y., Goncalves, A., Coin, L.J., Richardson, S., and Lewin, A. (2011). Haplotype and isoform specific expression estimation using multi-mapping RNA-seq reads. Genome Biol 12, R13.
Wang, L., Park, H.J., Dasari, S., Wang, S., Kocher, J.P., and Li, W. (2013). CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41, e74.
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10, 57–63.
Welter, D., MacArthur, J., Morales, J., Burdett, T., Hall, P., Junkins, H., Klemm, A., Flicek, P., Manolio, T., Hindorff, L., and Parkinson, H. (2014). The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nat Rev Genet 42, D1001–D1006.
Wu, A.R., Neff, N.F., Kalisky, T., Dalerba, P., Treutlein, B., Rothenberg, M.E., Mburu, F.M., Mantalas, G.L., Sim, S., Clarke, M.F., and Quake, S.R. (2014). Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods 11, 41–46.
Zhao, Y., Luo, H., Chen, X., Xiao, Y., and Chen, R. (2014). Computational methods to predict long noncoding RNA functions based on co-expression network. Methods Mol Biol 1182, 209–218.
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (2015AA020104), the China Human Proteome Project (2014DFB30010), the National Science Foundation of China (31471239, to Leming Shi), and the 111 Project (B13016).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, G., Shi, T. & Shi, L. Characterizing and annotating the genome using RNA-seq data. Sci. China Life Sci. 60, 116–125 (2017). https://doi.org/10.1007/s11427-015-0349-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-015-0349-4