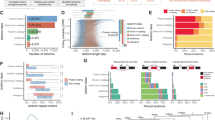
Abstract
Human and mouse orthologs are expected to have similar biological functions; however, many discrepancies have also been reported. We systematically compared human and mouse orthologs in terms of alternative splicing patterns and expression profiles. Human-mouse orthologs are divergent in alternative splicing, as human orthologs could generally encode more isoforms than their mouse orthologs. In early embryos, exon skipping is far more common with human orthologs, whereas constitutive exons are more prevalent with mouse orthologs. This may correlate with divergence in expression of splicing regulators. Orthologous expression similarities are different in distinct embryonic stages, with the highest in morula. Expression differences for orthologous transcription factor genes could play an important role in orthologous expression discordance. We further detected largely orthologous divergence in differential expression between distinct embryonic stages. Collectively, our study uncovers significant orthologous divergence from multiple aspects, which may result in functional differences and dynamics between human-mouse orthologs during embryonic development.
Article PDF
Similar content being viewed by others
References
Angel, T.E., Aryal, U.K., Hengel, S.M., Baker, E.S., Kelly, R.T., Robinson, E.W., and Smith, R.D. (2012). Mass spectrometry-based proteomics: existing capabilities and future directions. Chem Soc Rev 41, 3912–3928.
Blair, J.E., and Hedges, S.B. (2005). Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol 22, 2275–2284.
Chen, X., and Zhang, J. (2012). The ortholog conjecture is untestable by the current gene ontology but is supported by RNA sequencing data. PLoS Comput Biol 8, e1002784.
Ciccarelli, F.D., Doerks, T., von Mering, C., Creevey, C.J., Snel, B., and Bork, P. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287.
Dolinski, K., and Botstein, D. (2007). Orthology and functional conservation in eukaryotes. Annu Rev Genet 41, 465–507.
Domon, B., and Aebersold, R. (2006). Mass spectrometry and protein analysis. Science 312, 212–217.
Elso, C., Lu, X., Morrison, S., Tarver, A., Thompson, H., Thurkow, H., Yamada, N.A., and Stubbs, L. (2008). Germline translocations in mice: unique tools for analyzing gene function and long-distance regulatory mechanisms. J Natl Cancer Inst Monogr, 91–95.
Flicek, P., Ahmed, I., Amode, M.R., Barrell, D., Beal, K., Brent, S., Carvalho-Silva, D., Clapham, P., Coates, G., Fairley, S., Fitzgerald, S., Gil, L., Garcia-Giron, C., Gordon, L., Hourlier, T., Hunt, S., Juettemann, T., Kahari, A.K., Keenan, S., Komorowska, M., Kulesha, E., Longden, I., Maurel, T., McLaren, W.M., Muffato, M., Nag, R., Overduin, B., Pignatelli, M., Pritchard, B., Pritchard, E., Riat, H.S., Ritchie, G.R., Ruffier, M., Schuster, M., Sheppard, D., Sobral, D., Taylor, K., Thormann, A., Trevanion, S., White, S., Wilder, S.P., Aken, B.L., Birney, E., Cunningham, F., Dunham, I., Harrow, J., Herrero, J., Hubbard, T.J., Johnson, N., Kinsella, R., Parker, A., Spudich, G., Yates, A., Zadissa, A., and Searle, S.M. (2013). Ensembl 2013. Nucleic Acids Res 41, D48–55.
Gabaldon, T., and Koonin, E.V. (2013). Functional and evolutionary implications of gene orthology. Nat Rev Genet 14, 360–366.
Gharib, W.H., and Robinson-Rechavi, M. (2011). When orthologs diverge between human and mouse. Brief Bioinform 12, 436–441.
Ginis, I., Luo, Y.Q., Miura, T., Thies, S., Brandenberger, R., Gerecht-Nir, S., Amit, M., Hoke, A., Carpenter, M.K., Itskovitz-Eldor, J., and Rao, M.S. (2004). Differences between human and mouse embryonic stem cells. Dev Biol 269, 360–380.
Grosso, A.R., Gomes, A.Q., Barbosa-Morais, N.L., Caldeira, S., Thorne, N.P., Grech, G., von Lindern, M., and Carmo-Fonseca, M. (2008). Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res 36, 4823–4832.
Huang da, W., Sherman, B.T., and Lempicki, R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57.
Kanamori, M., Konno, H., Osato, N., Kawai, J., Hayashizaki, Y., and Suzuki, H. (2004). A genome-wide and nonredundant mouse transcription factor database. Biochem Biophys Res Commun 322, 787–793.
Keren, H., Lev-Maor, G., and Ast, G. (2010). Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11, 345–355.
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36.
Kinsella, R.J., Kahari, A., Haider, S., Zamora, J., Proctor, G., Spudich, G., Almeida-King, J., Staines, D., Derwent, P., Kerhornou, A., Kersey, P., and Flicek, P. (2011). Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011, bar030.
Koonin, E.V. (2005). Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39, 309–338.
Liao, B.Y., and Zhang, J. (2006). Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol Biol Evol 23, 530–540.
Liu, H., Chen, C.H., Espinoza-Lewis, R.A., Jiao, Z., Sheu, I., Hu, X., Lin, M., Zhang, Y., and Chen, Y. (2011). Functional redundancy between human SHOX and mouse Shox2 genes in the regulation of sinoatrial node formation and pacemaking function. J Biol Chem 286, 17029–17038.
Liu, Z., Miner, J.J., Yago, T., Yao, L., Lupu, F., Xia, L., and McEver, R.P. (2010). Differential regulation of human and murine P-selectin expression and function in vivo. J Exp Med 207, 2975–2987.
Marioni, J.C., Mason, C.E., Mane, S.M., Stephens, M., and Gilad, Y. (2008). RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18, 1509–1517.
McGettigan, P.A. (2013). Transcriptomics in the RNA-seq era. Curr Opin Chem Biol 17, 4–11.
Mouse Genome Sequencing, C., Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., Antonarakis, S.E., Attwood, J., Baertsch, R., Bailey, J., Barlow, K., Beck, S., Berry, E., Birren, B., Bloom, T., Bork, P., Botcherby, M., Bray, N., Brent, M.R., Brown, D.G., Brown, S.D., Bult, C., Burton, J., Butler, J., Campbell, R.D., Carninci, P., Cawley, S., Chiaromonte, F., Chinwalla, A.T., Church, D.M., Clamp, M., Clee, C., Collins, F.S., Cook, L.L., Copley, R.R., Coulson, A., Couronne, O., Cuff, J., Curwen, V., Cutts, T., Daly, M., David, R., Davies, J., Delehaunty, K.D., Deri, J., Dermitzakis, E.T., Dewey, C., Dickens, N.J., Diekhans, M., Dodge, S., Dubchak, I., Dunn, D.M., Eddy, S.R., Elnitski, L., Emes, R.D., Eswara, P., Eyras, E., Felsenfeld, A., Fewell, G.A., Flicek, P., Foley, K., Frankel, W.N., Fulton, L.A., Fulton, R.S., Furey, T.S., Gage, D., Gibbs, R.A., Glusman, G., Gnerre, S., Goldman, N., Goodstadt, L., Grafham, D., Graves, T.A., Green, E.D., Gregory, S., Guigo, R., Guyer, M., Hardison, R.C., Haussler, D., Hayashizaki, Y., Hillier, L.W., Hinrichs, A., Hlavina, W., Holzer, T., Hsu, F., Hua, A., Hubbard, T., Hunt, A., Jackson, I., Jaffe, D.B., Johnson, L.S., Jones, M., Jones, T.A., Joy, A., Kamal, M., Karlsson, E.K., Karolchik, D., Kasprzyk, A., Kawai, J., Keibler, E., Kells, C., Kent, W.J., Kirby, A., Kolbe, D.L., Korf, I., Kucherlapati, R.S., Kulbokas, E.J., Kulp, D., Landers, T., Leger, J.P., Leonard, S., Letunic, I., Levine, R., Li, J., Li, M., Lloyd, C., Lucas, S., Ma, B., Maglott, D.R., Mardis, E.R., Matthews, L., Mauceli, E., Mayer, J.H., McCarthy, M., McCombie, W.R., McLaren, S., McLay, K., McPherson, J.D., Meldrim, J., Meredith, B., Mesirov, J.P., Miller, W., Miner, T.L., Mongin, E., Montgomery, K.T., Morgan, M., Mott, R., Mullikin, J.C., Muzny, D.M., Nash, W.E., Nelson, J.O., Nhan, M.N., Nicol, R., Ning, Z., Nusbaum, C., O’Connor, M.J., Okazaki, Y., Oliver, K., Overton-Larty, E., Pachter, L., Parra, G., Pepin, K.H., Peterson, J., Pevzner, P., Plumb, R., Pohl, C.S., Poliakov, A., Ponce, T.C., Ponting, C.P., Potter, S., Quail, M., Reymond, A., Roe, B.A., Roskin, K.M., Rubin, E.M., Rust, A.G., Santos, R., Sapojnikov, V., Schultz, B., Schultz, J., Schwartz, M.S., Schwartz, S., Scott, C., Seaman, S., Searle, S., Sharpe, T., Sheridan, A., Shownkeen, R., Sims, S., Singer, J.B., Slater, G., Smit, A., Smith, D.R., Spencer, B., Stabenau, A., Stange-Thomann, N., Sugnet, C., Suyama, M., Tesler, G., Thompson, J., Torrents, D., Trevaskis, E., Tromp, J., Ucla, C., Ureta-Vidal, A., Vinson, J.P., Von Niederhausern, A.C., Wade, C.M., Wall, M., Weber, R.J., Weiss, R.B., Wendl, M.C., West, A.P., Wetterstrand, K., Wheeler, R., Whelan, S., Wierzbowski, J., Willey, D., Williams, S., Wilson, R.K., Winter, E., Worley, K.C., Wyman, D., Yang, S., Yang, S.P., Zdobnov, E.M., Zody, M.C., and Lander, E.S. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562.
Nagaraj, N., Wisniewski, J.R., Geiger, T., Cox, J., Kircher, M., Kelso, J., Paabo, S., and Mann, M. (2011). Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol 7, 548.
Nehrt, N.L., Clark, W.T., Radivojac, P., and Hahn, M.W. (2011). Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput Biol 7, e1002073.
Nurtdinov, R.N., Artamonova, II, Mironov, A.A., and Gelfand, M.S. (2003). Low conservation of alternative splicing patterns in the human and mouse genomes. Hum Mol Genet 12, 1313–1320.
Ozsolak, F., and Milos, P.M. (2011). RNA sequencing: advances, challenges and opportunities. Nature Rev Genet 12, 87–98.
Pal, S., Gupta, R., Kim, H., Wickramasinghe, P., Baubet, V., Showe, L.C., Dahmane, N., and Davuluri, R.V. (2011). Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res 21, 1260–1272.
Pereira, V., Waxman, D., and Eyre-Walker, A. (2009). A problem with the correlation coefficient as a measure of gene expression divergence. Genetics 183, 1597–1600.
Piasecka, B., Robinson-Rechavi, M., and Bergmann, S. (2012). Correcting for the bias due to expression specificity improves the estimation of constrained evolution of expression between mouse and human. Bioinformatics 28, 1865–1872.
Qian, W., Liao, B.Y., Chang, A.Y., and Zhang, J. (2010). Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet 26, 425–430.
Ramskold, D., Luo, S., Wang, Y.C., Li, R., Deng, Q., Faridani, O.R., Daniels, G.A., Khrebtukova, I., Loring, J.F., Laurent, L.C., Schroth, G.P., and Sandberg, R. (2012). Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 30, 777–782.
Rebhan, M., Chalifa-Caspi, V., Prilusky, J., and Lancet, D. (1998). Gene-Cards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 14, 656–664.
Roux, J., and Robinson-Rechavi, M. (2011). Age-dependent gain of alternative splice forms and biased duplication explain the relation between splicing and duplication. Genome Res 21, 357–363.
Shapiro, E., Biezuner, T., and Linnarsson, S. (2013). Single-cell sequencing- based technologies will revolutionize whole-organism science. Nat Rev Genet 14, 618–630.
Studer, R.A., and Robinson-Rechavi, M. (2009). How confident can we be that orthologs are similar, but paralogs differ? Trends Genet 25, 210–216.
Takeda, J., Suzuki, Y., Sakate, R., Sato, Y., Seki, M., Irie, T., Takeuchi, N., Ueda, T., Nakao, M., Sugano, S., Gojobori, T., and Imanishi, T. (2008). Low conservation and species-specific evolution of alternative splicing in humans and mice: comparative genomics analysis using well-annotated full-length cDNAs. Nucleic Acids Res 36, 6386–6395.
Trapnell, C., Hendrickson, D.G., Sauvageau, M., Goff, L., Rinn, J.L., and Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31, 46–53.
Trapnell, C., Williams, B.A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M.J., Salzberg, S.L., Wold, B.J., and Pachter, L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515.
van der Vegt, B., de Bock, G.H., Hollema, H., and Wesseling, J. (2009). Microarray methods to identify factors determining breast cancer progression: potentials, limitations, and challenges. Crit Rev Oncol Hematol 70, 1–11.
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10, 57–63.
Wingender, E., Schoeps, T., and Donitz, J. (2013). TFClass: an expandable hierarchical classification of human transcription factors. Nucleic Acids Res 41, D165–170.
Xing, Y., Ouyang, Z., Kapur, K., Scott, M.P., and Wong, W.H. (2007). Assessing the conservation of mammalian gene expression using high-density exon arrays. Mol Biol Evol 24, 1283–1285.
Xue, Z., Huang, K., Cai, C., Cai, L., Jiang, C.Y., Feng, Y., Liu, Z., Zeng, Q., Cheng, L., Sun, Y.E., Liu, J.Y., Horvath, S., and Fan, G. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597.
Yashiro, K., Saijoh, Y., Sakuma, R., Tada, M., Tomita, N., Amano, K., Matsuda, Y., Monden, M., Okada, S., and Hamada, H. (2000). Distinct transcriptional regulation and phylogenetic divergence of human LEFTY genes. Genes Cells 5, 343–357.
Yeo, G.W. (2005). Splicing regulators: targets and drugs. Genome Biol 6, 240.
Acknowledgements
We would like to thank Li Zhang and Rong Cheng from East China Normal University for their helpful discussions. We are also grateful to the Supercomputer Center of East China Normal University. This work was supported by the China Human Proteomics Project (2014DFB30010), the National High Technology Research and Development Program of China (2015AA020104,), the National Natural Science Foundation of China (31071162), and the Graduate School of East China Normal University.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
11427_2015_348_MOESM1_ESM.doc
Significant variations in alternative splicing patterns and expression profiles between human-mouse orthologs in early embryos
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, G., Chen, J., Yang, J. et al. Significant variations in alternative splicing patterns and expression profiles between human-mouse orthologs in early embryos. Sci. China Life Sci. 60, 178–188 (2017). https://doi.org/10.1007/s11427-015-0348-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-015-0348-5