Abstract
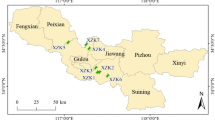
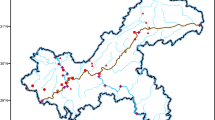
A survey was conducted in regions of North China to better understand the effect of antibiotic residue pollution from swine feedlots to nearby groundwater environment. A total of nine experimental sites located in the regions of Beijing, Hebei, and Tianjin were selected to analyze the presence of residues of 11 most commonly used antibiotics, including tetracyclines (TCs), fluoroquinolones (FQNs), sulfonamides (SAs), macrolides, and fenicols, by using liquid chromatography spectrometry. The three most common antibiotics were TCs, FQNs, and SAs, with mean concentrations of 416.4, 228.8, and 442.4 μg L−1 in wastewater samples; 19.9, 11.8, and 0.3 μg L−1 in groundwater samples from swine feedlots; and 29.7, 14.0, and 0 μg L−1 in groundwater samples from villages. Ordination analysis revealed that the composition and distribution of antibiotics and antibiotic resistance genes (AGRs) were similar in groundwater samples from swine feedlots and villages. FQNs and TCs occurred along the path from wastewater to groundwater at high concentrations and showed correlations with ARGs, with a strong correlation between FQN resistance gene (qnrA) copy number. FQN concentration was also found (P < 0.01) in wastewater and groundwater in villages (P < 0.01). Therefore, antibiotics discharged from swine feedlots through wastewater could disseminate into surrounding groundwater environments together with ARG occurrence (i.e., qnrA, sulI, sulII, tetG, tetM, and tetO). Overall, this study suggests that the spread of veterinary antibiotics from swine feedlots to groundwater environments should be highly attended and controlled by restricting excess antibiotic usage or improving the technology of manure management.





Similar content being viewed by others
References
Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8(4):251–259. https://doi.org/10.1038/nrmicro2312
Aminov RI, Garrigues-Jeanjean N, Mackie RI (2001) Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol 67(1):22–32
Apley MD, Bush EJ, Morrison RB (2012) Use estimates of in-feed antimicrobials in swine production in the United States. Foodborne Pathog Dis 9(3):272–279. https://doi.org/10.1089/fpd.2011.0983
Binh CTT, Heuer H, Kaupenjohann M, Smalla K (2008) Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol Ecol 66(1):25–37. https://doi.org/10.1111/j.1574-6941.2008.00526.x
Brooks JP, Adeli A, McLaughlin MR (2014) Microbial ecology, bacterial pathogens, and antibiotic resistant genes in swine manure wastewater as influenced by three swine management systems. Water Res 57:96–103
Chen K, Zhou JL (2014) Occurrence and behavior of antibiotics in water and sediments from the Huangpu River, Shanghai, China. Chemosphere 95:604–612. https://doi.org/10.1016/j.chemosphere.2013.09.119
Chinese Ministry of Agriculture (2016) Yearbook of animal husbandry and veterinary medicine of China
Czekalski N, Berthold T, Caucci S, Egli A, Bürgmann H (2012) Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into lake Geneva, Switzerland. Front Microbiol 3:106
Feng Y, Wei C, Zhang W, Liu Y, Li Z, Hu H, Xue J, Davis M (2016) A simple and economic method for simultaneous determination of 11 antibiotics in manure by solid-phase extraction and high-performance liquid chromatography. J Soils Sediments 16(9):2242–2251. https://doi.org/10.1007/s11368-016-1414-5
Gao P, Munir M, Xagoraraki I (2012) Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci Total Environ 421–422:173–183
Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M (2008) The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol 190(14):5095–5100. https://doi.org/10.1128/JB.00152-08
Heuer H, Schmitt H, Smalla K (2011) Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol 14:236–243
Hirsch R, Ternes T, Haberer K, Kratz K (1999) Occurrence of antibiotics in the aquatic environment. Sci Total Environ 225(1-2):109–118. https://doi.org/10.1016/S0048-9697(98)00337-4
Hu XG, Luo Y, Zhou QX, Xu L (2008) Determination of thirteen antibiotics residues in manure by solid phase extraction and high performance liquid chromatography. Chin J Anal Chem 38:1162–1166
Jiang H, Zhang D, Xiao S, Geng C, Zhang X (2013) Occurrence and sources of antibiotics and their metabolites in river water, WWTPs, and swine wastewater in Jiulongjiang River basin, south China. Environ Sci Pollut Res Int 20:9075–9083
Kümmerer K (2009) Antibiotics in the aquatic environment—a review—part II. Chemosphere 75(4):435–441. https://doi.org/10.1016/j.chemosphere.2008.12.006
Lepx J, Smilauer P (2003) Multivariate analysis of ecological data using CANOCO. Cambridge University Press, Cambridge, pp 50–52. https://doi.org/10.1017/CBO9780511615146
Liu Y, Guan YT, Gao BY, Yue QY (2012) Antioxidant responses and degradation of two antibiotic contaminants in Microcystis aeruginosa. Ecotoxicol Environ Saf 86:23–30. https://doi.org/10.1016/j.ecoenv.2012.09.004
Ma YP, Li M, Wu MM, Li Z, Liu X (2015) Occurrences and regional distributions of 20 antibiotics in water bodies during groundwater recharge. Sci Total Environ 518–519:498–506
Manzetti S, Ghisi R (2014) The environmental release and fate of antibiotics. Mar Pollut Bull 79(1-2):7–15. https://doi.org/10.1016/j.marpolbul.2014.01.005
Mazel D (2006) Integrons: agents of bacterial evolution. Nat Rev Microbiol 4(8):608–620. https://doi.org/10.1038/nrmicro1462
Mckinney CW, Loftin KA, Meyer MT, Davis JG, Pruden A (2010) Tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ Sci Technol 44(16):6102–6109. https://doi.org/10.1021/es9038165
Negreanu Y, Pasternak Z, Jurkevitch E, Cytryn E (2012) Impact of treated wastewater irrigation on antibiotic resistance in agricultural soils. Environ Sci Technol 46(9):4800–4808. https://doi.org/10.1021/es204665b
Pei R, Kim SC, Carlson KH, Pruden A (2006) Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40(12):2427–2435. https://doi.org/10.1016/j.watres.2006.04.017
Rodriguez-Mozaz S, Chamorro S, Marti E, Huerta B, Gros M, Sanchez-Melsio A, Borrego CM, Barcelo D, Balcazar JL (2015) Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res 69:234–242
Sacher F, Lang FT, Brauch HJ, Blankenhorn I (2001) Pharmaceuticals in groundwaters — analytical methods and results of a monitoring program in Baden-Wurttemberg, Germany. J Chromatogr A 938(1-2):199–210
Sentchilo V, Mayer AP, Guy L, Miyazaki R, Tringe SG, Barry K, Malfatti S, Goessmann A, Robinson-Rechavi M, van der Meer JR (2013) Community-wide plasmid gene mobilization and selection. ISME J 7(6):1173–1186. https://doi.org/10.1038/ismej.2013.13
Stokes HW, Nesbø CL, Holley M, Bahl MI, Gillings MR, Boucher Y (2006) Class 1 integrons potentially predating the association with tn402-like transposition genes are present in a sediment microbial community. J Bacteriol 188(16):5722–5730. https://doi.org/10.1128/JB.01950-05
Storey JM, Clark SB, Johnson AS, AndersenWC TSB, Lohne JJ, Burger RJ, Ayres PR, Carr JR, Madson MR (2014) Analysis of sulfonamides, trimethoprim, fluoroquinolones, quinolones, triphenylmethane dyes and methyltestosterone in fish and shrimp using liquid chromatography-mass spectrometry. J Chromatogr B 972:38–47. https://doi.org/10.1016/j.jchromb.2014.09.009
Tao CW, Hsu BM, Ji WT, Hsu TK, Kao PM, Hsu CP, Shen SM, Shen TY, Wan TJ, Huang YL (2014) Evaluation of five antibiotic resistance genes in wastewater treatment systems of swine farms by real-time PCR. Sci Total Environ 496:116–121. https://doi.org/10.1016/j.scitotenv.2014.07.024
Thomas KV, Dye C, Schlabach M, Langford KH (2007) Source to sink tracking of selected human pharmaceuticals from two Oslo city hospitals and a wastewater treatment works. J Environ Monit 9(12):1410–1418. https://doi.org/10.1039/b709745j
Wang J, Ben W, Yang M, Zhang Y, Qiang Z (2016) Dissemination of veterinary antibiotics and corresponding resistance genes from a concentrated swine feedlot along the waste treatment paths. Environ Inter 92–93:317–323
Xu J, Xu Y, Wang H, Guo C, Qiu H, He Y, Zhang Y, Li X, Meng W (2015) Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 119:1379–1385. https://doi.org/10.1016/j.chemosphere.2014.02.040
Zhang T, Zhang XX, Ye L (2011) Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS One 6(10):e26041. https://doi.org/10.1371/journal.pone.0026041
Zhao S, Liu X, Cheng D, Liu G, Liang B, Cui B, Bai J (2016) Temporal-spatial variation and partitioning prediction of antibiotics in surface water and sediments from the intertidal zones of the Yellow River Delta, China. Sci Total Environ 569–570:1350–1358
Zhao X, Wang J, Zhu L, Ge W, Wang J (2017) Environmental analysis of typical antibiotic-resistant bacteria and ARGs in farmland soil chronically fertilized with chicken manure. Sci Total Environ 593–594:10–17
Zhou LJ, Ying GG, Liu S, Zhang RQ, Lai HJ, Chen ZF, Pan CG (2013) Excretion masses and environmental occurrence of antibiotics in typical swine and dairy cattle farms in China. Sci Total Environ 44:183–195
Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM (2013) Diverse and abundant antibiotic resistance genes in Chinese swine farms. PNAS 110(9):3435–3440. https://doi.org/10.1073/pnas.1222743110
Acknowledgments
This study was financially supported by the Science and Technology Program of Tianjin (14ZCZDNC00013) and the National Key Research and Development Program of China (2016YFD080060 and 2017YFD0801400).
Author information
Authors and Affiliations
Additional information
Responsible editor: Ester Heath
Xiaohua Li and Chong Liu were our first co-authors
Rights and permissions
About this article
Cite this article
Li, X., Liu, C., Chen, Y. et al. Antibiotic residues in liquid manure from swine feedlot and their effects on nearby groundwater in regions of North China. Environ Sci Pollut Res 25, 11565–11575 (2018). https://doi.org/10.1007/s11356-018-1339-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-018-1339-1