Abstract
Purpose
Dynamic positron emission tomography (PET) protocols allow for accurate quantification of [18F]flortaucipir-specific binding. However, dynamic acquisitions can be challenging given the long required scan duration of 130 min. The current study assessed the effect of shorter scan protocols for [18F]flortaucipir on its quantitative accuracy.
Procedures
Two study cohorts with Alzheimer’s disease (AD) patients and healthy controls (HC) were included. All subjects underwent a 130-min dynamic [18F]flortaucipir PET scan consisting of two parts (0–60/80–130 min) post-injection. Arterial sampling was acquired during scanning of the first cohort only. For the second cohort, a second PET scan was acquired within 1–4 weeks of the first PET scan to assess test-retest repeatability (TRT). Three alternative time intervals were explored for the second part of the scan: 80–120, 80–110 and 80–100 min. Furthermore, the first part of the scan was also varied: 0–50, 0–40 and 0–30 min time intervals were assessed. The gap in the reference TACs was interpolated using four different interpolation methods: population-based input function 2T4k_VB (POP-IP_2T4k_VB), cubic, linear and exponential. Regional binding potential (BPND) and relative tracer delivery (R1) values estimated using simplified reference tissue model (SRTM) and/or receptor parametric mapping (RPM). The different scan protocols were compared to the respective values estimated using the original scan acquisition. In addition, TRT of the RPM BPND and R1 values estimated using the optimal shortest scan duration was also assessed.
Results
RPM BPND and R1 obtained using 0–30/80–100 min scan and POP-IP_2T4k_VB reference region interpolation had an excellent correlation with the respective parametric values estimated using the original scan duration (r2 > 0.95). The TRT of RPM BPND and R1 using the shortest scan duration was − 1 ± 5 % and − 1 ± 6 % respectively.
Conclusions
This study demonstrated that [18F]flortaucipir PET scan can be acquired with sufficient quantitative accuracy using only 50 min of dual-time-window scanning time.
Similar content being viewed by others
Introduction
Dynamic positron emission tomography (PET) scan protocols allow for accurate quantitative measures [1, 2] of specific binding of PET tracers. Moreover, dynamic scan protocols yield additional information about functional measures such as perfusion [3]. Semi-quantitative measures from static scans are usually sufficient for clinical application, but accurate quantification of tracer uptake is of major importance in the context of early-stage pathology, clinical trials [1] and longitudinal studies. Some PET tracers like the tau tracer [18F]flortaucipir require a long acquisition period because of the slow tracer kinetics. This can be challenging, especially when working with a vulnerable population (like patients with Alzheimer’s disease (AD)).
In vivo quantification of tau pathology is important because intracellular accumulation of hyperphosphorylated tau proteins into neurofibrillary tangles (NFTs) is one of the pathological hallmarks of AD [4]. Indeed, histopathological studies have shown that the amount of NFTs correlate well with the severity of their cognitive symptoms during life [5, 6]. [18F]Flortaucipir is worldwide the most widely used PET tracer for detecting and quantifying these NFTs. For the analysis of [18F]flortaucipir scans, most studies prefer semi-quantitative measures due to their practical applicability and computational simplicity [7,8,9]. However, studies involving dynamic imaging provided more accurate and precise pharmacokinetic parameters and provide estimates for relative tracer delivery (R1) or relative cerebral blood flow (rCBF) [2, 10,11,12,13,14,15], which is important for monitoring flow changes. For instance, a study by van Berckel et al. [16] observed that longitudinal changes in [11C]PIB standardized uptake value ratio (SUVr) do not reflect changes in specific [11C]PIB binding but rather are secondary to changes in blood flow during the natural course of AD.
Our group has performed dynamic acquisition of [18F]flortaucipir scans, using a 130-min dual-time-window dynamic scan protocol including a 20-min break (after the first 60 min of acquisition) [17,18,19,20,21]. Several aspects are of importance to obtain a reliable protocol with reduced overall scanning time. Firstly, the scan must include the wash-in of the tracer and tissue peak activity to be able to assess the tracer influx into the tissue. In addition, tracer efflux information is also necessary to be able to estimate the tracer efflux back to plasma and the specific binding compartment. The second part ideally has to contain the 80–100 min interval to calculate SUVr, since this is the internationally conventional SUVr interval for [18F]flortaucipir [22]. So, the new scanning protocol needs to include an early part of the tracer kinetics and also at least 80–100 min post-injection (p.i.), implying that a dual-time-window protocol should be used. Scanning time can be shortened by increasing the gap of the dual-time-window. Interpolation is needed to fill this gap in the time activity curve (TAC) of the reference region to be able to perform reference tissue model–based tracer kinetic modelling. Therefore, the aim of the study is to investigate whether a shorter overall scan duration for [18F]flortaucipir PET dual-time-window scans is feasible, while retaining quantitative accuracy.
Methods
Study Sample
For the current project, two study cohorts were included. The first cohort consisted of ten biomarker (PET/CSF)-confirmed AD patients and ten cognitively normal controls who underwent a 130-min dynamic [18F]flortaucipir PET scan with arterial sampling (“full kinetic model cohort”). Subject characteristics have been described previously [18]. The second cohort consisted of eight subjects with AD and six cognitively normal controls that underwent two 130-min dynamic [18F]flortaucipir PET scans within a time interval of minimum 1 week, and maximum 4 weeks (“test-retest cohort”). The subject characteristics have been described previously [19]. The current study was approved by the Medical Ethics Committee of the Amsterdam University Medical Center. All subjects signed an informed consent form prior to participation.
Scan Procedures
T1-weighted MRI scans were acquired for all participants using a 3.0 T Philips Ingenuity Time-of-Flight PET/MR scanner (Philips medical systems, Best, the Netherlands). Isotropic structural 3D T1-weighted images were obtained using a sagittal turbo field echo sequence (1.00 mm3 isotropic voxels, repetition time = 7.9 ms, echo time = 4.5 ms, flip angle = 8°) for brain tissue segmentation.
All subjects from the full kinetic model cohort underwent a 130-min dynamic [18F]flortaucipir PET scan on a Gemini TF-64 PET/CT scanner (Philips Medical Systems, Best, The Netherlands) with continuous arterial sampling after administration of 223 ± 18 MBq of [18F]flortaucipir. Details described elsewhere [17,18,19]. Subjects from the test-retest cohort underwent two 130-min dynamic [18F]flortaucipir PET scans on a Philips Ingenuity TF PET/CT scanner after administration of [18F]flortaucipir (237 ± 15 MBq at test and 245 ± 18 MBq at retest) as described in detail previously [19]. In short, a low-dose CT for attenuation correction was acquired, followed by a 60-min dynamic (brain) emission scan initiated simultaneously with tracer injection. After a 20-min break, a second low-dose CT was acquired before an additional dynamic emission scan during the interval 80–130 min p.i. During scanning, the head of the subjects was stabilized to reduce movement artefacts. Furthermore, subjects were positioned within the centre of axial and transaxial fields of view, such that the orbitomeatal line was parallel to the detectors with the use of laser beams.
For the full kinetic model cohort, continuous arterial blood sampling, using an online detection, [23] was collected during 60-min p.i. PET acquisition. Furthermore, manual arterial samples were collected at set time points (5, 10, 15, 20, 40, 60, 80, 105 and 130 min p.i.) to measure plasma metabolite fractions and plasma-to-whole-blood ratios. Using the aforementioned information, the continuous online blood sampler data was calibrated and corrected for metabolites, plasma-to-whole-blood ratios and delay, providing a metabolite-corrected arterial plasma input function. In addition, whole-blood input function was obtained for blood volume correction.
Image Processing
PET scans were reconstructed with a matrix size of 128 × 128 × 90 and a final voxel size of 2 × 2 × 2 mm3. All standard corrections were applied. During processing of the PET scans, first part and second part of the scan were checked for motion, separately. Thereafter, both the PET scan sessions were co-registered into a single dataset of 29 frames (1 × 15, 3 × 5, 3 × 10, 4 × 60, 2 × 150, 2 × 300, 4 × 600 and 10 × 300 s) using Vinci software (Max Plank Institute, Cologne, Germany). The last 10 frames belonged to the second PET session.
Structural 3D T1-weighted MRI images were co-registered to the PET images also using Vinci software (Max Plank Institute, Cologne, Germany). The Hammers template [24], which is incorporated in PVElab [25], was used to delineate regions of interest (ROIs) on the co-registered MR scan and superimposed onto the dynamic PET scan to obtain regional time activity curves (TACs). All 68 cortical and subcortical regions from the Hammer template were included. Regional TACs extracted from the PET scans were analysed using a reversible 2-tissue compartment model with blood volume correction (2T4k_VB) and simplified reference tissue model (SRTM) [26]. Receptor parametric mapping (RPM) [27] and standardized uptake value ratios (SUVr) were used to obtain parametric images. Cerebellum grey matter (obtained from PVElab) was used as the reference region.
Shortening the Second Part of the Scan (80–130 Min P.I.)
In these analyses, the first part of the scan remained 0–60 min p.i. The second part of the scan was shortened; three shorter time intervals were explored: 80–120 min, 80–110 min and 80–100 min. For each subject, shortened PET scans were acquired by removing 2 to 6 frames to reach the specified scan intervals. Reference region TACs were extracted from these shortened PET scans to estimate kinetic parameters. BPND and R1 values were estimated using RPM from the three different scan durations (0–60/80–100, 0–60/80–110 and 0–60/80–120 min). RPM-derived regional BPND and R1 values were compared to the corresponding non-linear regression (NLR)-based SRTM-derived BPND and R1, and plasma input–derived distribution volume ratio (DVR) values from the original scan duration (0–60/80–130 min). The optimal shortened time interval for the second part was used and fixed during subsequent evaluation of scan shortening of the first part of the PET scan.
Shortening the First Part of the Scan (0–60 Min P.I.)
For shortening the first part of the scan, three time intervals were explored: 0–50, 0–40 and 0–30 min p.i, all in combination with 80–100 min scan interval for the second part of the imaging protocol. For each subject, the corresponding frames were removed to obtain the PET scans with these specified time intervals.
The original scan duration had a gap of only 20 min; the gap in the reference region was interpolated by using cubic interpolation. The larger gap (> 20 min) in the new dual-time-window protocol results in more missing data points in the reference TAC for which proper interpolation is required. Therefore, four different interpolation methods were assessed: population-based plasma input function in combination with a reversible two-tissue compartmental model with blood volume correction (POP-IP_2T4k_VB) to fit the reference tissue TAC, standard cubic interpolation, linear interpolation, and interpolation based on fitting an exponential function to the TAC (excluding points until peak uptake). All scripts were built in house using MATLAB (version R2017B, MathWorks, USA).
The POP-IP_2T4K_VB interpolation method was based on using the population-averaged metabolite-corrected plasma input function and a reversible two-tissue compartmental model with blood volume correction (2T4k_VB). A 2T4k_VB model was used, since it was evaluated in the previous studies [28] that this model best describes the in vivo kinetics of [18F]flortaucipir. So based on the previous research, it was assumed that the cerebellum presents a 2T4k_VB kinetics and the cerebellum TAC with the gap was fitted using this model and the population-averaged metabolite-corrected plasma input function. The fit was visually examined for certainty and the gap in the cerebellum TAC was filled using the values from the fitted curve.
SRTM-derived BPND and R1 estimates using the shortened scan durations and the four different interpolated reference region TACs were obtained. These regional parametric values were compared to the corresponding NLR-based reference region and plasma input–derived values obtained using the original scan duration (0–60/80–130 min).
BPND and R1 parametric images were acquired for the optimally shortened scans with interpolated reference region (using the optimal interpolation technique(s)). Regional parametric values were extracted from these parametric images and were compared to corresponding values derived using plasma input–based and reference tissue–based NLR and RPM from the original scan duration (0–60/80–130 min).
SUVr using the interval 80–100 min (SUVr(80−100 min)) was also evaluated. Regional SUVr values obtained from this time interval were compared with the respective quantitative parameters (DVR, SRTM BPND and RPM BPND) estimated using the original scan duration (0–60/80–130 min).
Test-Retest Repeatability Analysis
For the test-retest repeatability (TRT) analysis, the test-retest cohort was used. The TRT of RPM BPND and R1 values derived from the optimal shortened scan duration were compared to the test-retest repeatability of RPM BPND obtained using the original scan duration (0–60/80–130 min). In addition, TRT for SUVr(80–100) was also assessed. The TRT was calculated using Eq. 1.
Statistical Analysis
Linear regression fitting and correlation coefficients (r2) were used to compare BPND and R1 for the shortened scan durations and SUVr(80−100 min) against corresponding parametric values for the original scan duration (0–60/80–130 min) derived from plasma input–based and reference tissue–based NLR and RPM. Furthermore, Bland-Altman plots were used to assess and illustrate TRT performance.
Results
Shortening the Second Part of the Scan (80–130 Min P.I.)
The RPM BPND values obtained from the three shortened scan durations (0–60/80–120, 0–60/80–110 and 0–60/80–100 min) provided excellent correlations with plasma input DVR-1, SRTM BPND and RPM BPND obtained using the original acquisition time window (0–60/80–130) (Table 1; all r2 > 0.93). Reduction of the time interval of the second part to 100 min had negligible effects on the RPM BPND estimation: correspondence to DVR-1 (HC: r2 = 0.94, slope = 0.95; AD: r2 = 0.94, slope = 0.92), SRTM BPND (HC: r2 = 0.98, slope = 1.05; AD: r2 = 0.96, slope = 0.85) and RPM BPND (HC: r2 = 0.98, slope = 1.04; AD: r2 = 0.99, slope = 0.94). Comparison of regional SRTM BPND values obtained using the shorter time interval (0–60/80–100) to plasma input DVR-1 and SRTM BPND obtained with the original scan duration (0–60/80–130) are presented in Supplementary Table 1.
The RPM R1 values obtained from the three shortened scan durations (0–60/80–120, 0–60/80–110 and 0–60/80–100) also provided excellent correlations with SRTM R1 and RPM R1 estimated using the original acquisition time window (0–60/80–130) (Supplementary Table 2).
Shortening the First Part of the Scan (0–60 Min P.I.)
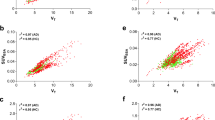
In Fig. 1, the different interpolations of a typical reference TAC for the shortest dual-time-window (0–30/80–100 min) assessed in this study are presented. For all shortened scan durations, SRTM BPND using the reference TACs interpolated with either POP-IP_2T4k_VB or cubic interpolation methods had the best correspondence with plasma input DVR-1 and SRTM BPND (r2 > 0.90, Table 2) obtained with the original scan duration. Reduction of the time interval of the first part of the scan to 30 min and using POP-IP_2T4k_VB for reference region interpolation had negligible effects on the quantitative accuracy of the estimated kinetic parameters with respect to that estimated using the original scan duration: DVR-1 (HC: r2 = 0.93, slope = 0.94; AD: r2 = 0.92, slope = 0.97) and SRTM BPND (HC: r2 = 0.96, slope = 1.02; AD: r2 = 0.98, slope = 0.89). SRTM BPND values obtained with 0–30/80–100 min data using cubic interpolation for reference region had similar agreement with DVR-1 (HC: r2 = 0.94, slope = 0.91; AD: r2 = 0.91, slope = 0.92) and SRTM BPND (HC: r2 = 0.96, slope = 0.98; AD: r2 = 0.98, slope = 0.85) from the original scan duration. Good correlations were observed for linear and exponential interpolation methods (r2 > 0.90, Table 2). However, these interpolation methods resulted in higher underestimation (15–25 %) of the parametric values.
Figure 2 presents the correspondence of SRTM BPND values obtained with the shortened scan durations using POP-IP_2T4k_VB for reference region interpolation against SRTM BPND values estimated from the original scan duration. The bias increased as the first part was shortened. An underestimation of 9 % was observed for SRTM BPND values with the shortened scan duration for both groups (0–30/80–100 min) with respect to that obtained with original scan duration. SRTM R1 values derived from the shortened scan duration (0–30/80–100 min) showed excellent correlations with SRTM and RPM R1 values obtained with the original scan duration for HC and AD patients for each interpolation method (r2 > 0.95, Supplementary Table 3).
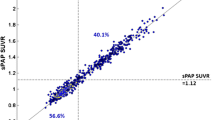
An example of the RPM BPND images for the original scan duration and shortened scan duration (0–30/80–100) is illustrated in Fig. 3a. Comparison of RPM BPND obtained from the shortest scan duration (0–30/80–100 min) using POP-IP_2T4k_VB for reference region interpolation against RPM BPND obtained with the original scan duration is shown in Fig. 3b and Supplementary Fig. 1. RPM BPND obtained with the shortest scan duration (0–30/80–100 min) using either POP-IP_2T4k_VB or cubic methods for reference region interpolation and SUVr80-100 min compared to original DVR-1, SRTM BPND and RPM BPND values are shown in Table 3 for HC and AD patients separately. The same comparisons were made for RPM R1 and are illustrated in Supplementary Table 4.
a An example of the BPND images for a representative AD patient are displayed for the original scan (0–60/80–130 min) duration and shortened scan duration (0–30/80–100 min using POP-IP_2T4k_VB interpolation) along with the corresponding MR. b Comparison of BPND obtained from the shortened scan duration (0–30/80–100 min using POP-IP_2T4k_VB interpolation) against BPND obtained with the original scan duration (0–60/80–130 min). c Bland-Altman plot of the original test-retest differences for RPM DVR (BPND+1) values. d Bland-Altman plot of the test-retest differences for RPM DVR (BPND+1) values using shortened scan duration (0–30/80–100 min) and POP-IP_2T4k_VB method for reference region interpolation.
Test-Retest Repeatability
The RPM BPND values estimated for both test and retest scans using the shortest assessed scan duration (0–30/80–100) and POP-IP_2T4k_VB or cubic interpolation for the reference region correlated well (Table 4). The test-retest parametric correlations using the short scan duration and interpolated reference TAC (POP-IP_2T4k_VB and cubic interpolation methods) were similar to the correlations using the original scan duration. The TRT across all Hammers’ regions of interest for RPM BPND was − 1 % ± 4 for HC and 0 % ± 4 for AD patients using the original dual-time-window acquisition (Table 4). Furthermore, TRT for R1 was 0 % ± 6 for both groups using the original dual-time-window acquisition. The TRT for BPND remained the same for HC when using the shortest dual-time-window 0–30/80–100 min with either of the interpolations methods (POP-IP_2T4k_VB or cubic interpolation). For AD patients, the TRT changed to 0 % ± 5 when using the shortest dual-time-window 0–30/80–100 min with POP-IP_2T4k_VB or cubic interpolation. The TRT for R1 remained similar as with the original data (0 % ± 6) for the shortest dual-time-window (0–30/80–100 min) both with POP-IP_2T4k_VB and cubic interpolation for both groups. TRT for SUVr80-100 min was − 1 % ± 5 for HC and − 1 % ± 6 for AD patients (Table 4).
Bland-Altman plots for the RPM BPND+1 illustrating TRT for all Hammers’ regions obtained with the original scan duration and the shortest scan duration with POP-IP_2T4k_VB reference region interpolation are presented in Fig. 3c and d.
Discussion
The current study demonstrated that for [18F]flortaucipir expanding the break in the dual-time-window protocol with just a 50-min overall scanning time (early interval of 0–30 min, than a coffee break, followed by a late interval of 80–100 min) had minimal effect on the quantitative accuracy. The optimal shortened dual-time-window protocol (0–30/80–100 min) allows sufficiently accurate estimation of BPND while reducing patient burden and enables interleaved scanning, where other patients could use the camera during breaks within the scan period.
An excellent correlation was observed between the shortened acquisition protocol (0–30/80–100 min) and the original dual-time-window (0–60/80–130 min) protocol. Four different interpolation methods were used to interpolate the missing data between the two time windows for the reference region TAC (cerebellum grey matter). According to our results, POP-IP_2T4k_VB interpolation, which uses a population-averaged plasma input function, showed a good correspondence of the estimated kinetic parameters to that obtained from the original scan protocol, and the lowest under/over-estimation(s) compared to other interpolation methods. Heeman et al. [29] showed that POP-IP_2T4k_VB interpolation method also works well to interpolate the missing data points in a dual-time-window protocol for [18F]flutemetamol and [18F]florbetaben. They concluded that the introduction of a gap with a maximum of 60 min in a dual-time-window protocol (early interval of 0–30 min followed by a late interval of 90–110 min) does not affect quantitative accuracy for [18F]flutemetamol and [18F]florbetaben. As such, POP-IP_2T4k_VB interpolation does not only work well for [18F]flortaucipir but also for [18F]flutemetamol and [18F]florbetaben, possibly because the model describes the in vivo kinetics of the tracers best and is therefore ideal for estimating the missing reference region data points accurately.
The correlations for all interpolation methods were comparable (Table 2). This was not expected, since linear and exponential interpolation did not follow the course of tracer as can be observed in Fig. 1. A possible explanation could be that the gap between the dual-time-windows was not substantially large enough to see significant differences in correlations between the interpolation methods. However, a substantial underestimation (at times up to 20 % or even more) was observed in AD patients for the shortened SRTM BPND values obtained with linear and exponential interpolation methods when compared to plasma input DVR-1 and SRTM BPND obtained with the original scan duration. This indicates that these interpolation methods are not suitable for quantitatively accurate kinetic parameter estimation for [18F]flortaucipir. For SRTM BPND values obtained with POP-IP_2T4k_VB interpolation, the biases remained within ~ 10 % for the same comparisons (Table 2). Comparisons of RPM BPND obtained with the shortest scanning interval (0–30/80–100) to plasma input DVR-1, SRTM BPND and RPM BPND derived from the original scan duration (0–60/80–130) showed that POP-IP_2T4k_VB interpolation had a minimal and acceptable bias of ~ 5 % (Table 3). Since RPM BPND is the main parameter outcome for [18F]flortaucipir, shortening the scan duration to 0–30/80–100 min with POP-IP_2T4k_VB interpolation for the reference region will provide quantitative acceptable accurate results. However, when individual regions are assessed, relatively higher bias (~ 8 %) was observed in regions with higher tau uptake (RPM DVR > 2) (Supplementary Fig. 1).
From Table 3, it is evident that SUVr(80-100 min) presents over-/underestimations when compared to DVR-1, SRTM BPND and RPM BPND (using 0–60/80–130 min scan duration data). In contrast, parameters estimated using a shortened scan duration data had a much better correspondence with the parameters estimated using the whole scan duration data (0–60/80–130 min). Although with a shortened scan duration, a 50-min scanning time is required, which is 30 min more than that required for a static scan; SUVr is still semi-quantitative. Moreover, it is known that SUVr might be effected by blood flow changes overtime, and hence is an unreliable parameter for longitudinal studies, whereas using the shortened dual-time-window protocol (0–30/80–100), a flow estimate (R1) to evaluate the effect of flow can be estimated and therefore is apt for longitudinal studies. Moreover with the proposed method, only 50 min of actual scanning time is required on contrast to 110 min of actual scan duration (decreasing the patient burden by 60 min of scanning).
Dynamic [18F]flortaucipir PET scans can be used to estimate both the specific binding of the tracer to tau pathology (BPND) and as well as rCBF (R1) using RPM. Perfusion imaging is a reliable biomarker to assess neuronal dysfunction in neurodegenerative diseases [30]. R1 is an estimate of the relative blood flow, and it has been shown that it has a high correlation with [18F]FDG uptake [30] and so an accurate estimation of R1 is also necessary. The current study demonstrated that shortening the scan duration to 0–30/80–100 min had negligible effects on R1 estimations. As such, it can be safely assumed that the scanning protocol can be reduced to 0–30/80–100 min with minimal bias (~ 7 % on average). If the implementation for POP-IP_2T4k_VB interpolation is not possible, use of cubic interpolation is a reliable alternative to interpolate the gap between the dual-time-windows for the reference region.
Unfortunately, further reduction of the acquisition time for the first part was not possible since the bias in the estimated parameters was increasing as the scan duration was being reduced (Fig. 2). A reason could be that as the peak approaches, further loss of information makes it difficult to estimate the efflux kinetics of [18F]flortaucipir. Further reduction of the second part of the scan was also not possible, since we want to be able to compare with the internationally mostly used 80–100-min SUVr time interval.
The shortened dual-time-window (0–30/80–100 min) acquisition protocol provided almost identical TRT values when compared to the observed TRT values for the original scan protocol (0–60/80–130) (Fig. 3). This implies that the shortened dual-time-window protocol not only provides quantitatively acceptable estimates but also result in high repeatability, suggesting that it can be reliably used for longitudinal and treatment-monitoring studies. However, the benefits of using a dynamic scanning protocol over static scanning protocol in a longitudinal setup for [18F]Flortaucipir need further validation, even though it has been presented by van Berckel et al. [16] that SUVr does get affected by changes in flow and under-/overestimates the underlying specific binding in case of [11C]PIB. Making use of a dual-time-window protocol also has some challenges to consider. Even if the total acquisition time is reduced with 60 min, it still takes longer than a single static acquisition protocol used for SUVr. Another disadvantage of using a dual-time-window protocol is the added CT attenuation scan for the second part of the scan, which is still present in this new shortened scanning protocol. Furthermore, the pre-processing of the PET images are more demanding compared to simplified methods, for instance, due to a required additional co-registration of the first part to the second part of the scan. Yet, the main advantage is that, apart from obtaining quantitative information on BPND, this protocol also generates parametric R1 data that may be used as a surrogate for flow and/or FDG uptake [15] and this protocol could therefore obviate the need to make a separate FDG scan, when clinically required.
Conclusion
The current study demonstrated that quantitatively acceptable [18F]flortaucipir kinetic parameters can be obtained using just 50 min of total scanning time by implementing a dual-time-window protocol (0–30/80–100 min). The best method to interpolate the gap in the reference tissue is 2T4k_VB tracer kinetic model with population-averaged metabolite-corrected plasma input function. Reducing the dual-time-window protocol enables interleaved scanning, reduces patient burden and enables efficient use of tracer batches and cost-effectiveness.
References
Lammertsma AA (2017) Forward to the past: the case for quantitative PET imaging. J Nucl Med 58:1019–1024
Ossenkoppele R, Prins ND and van Berckel BN. Amyloid imaging in clinical trials. Alzheimer's research & therapy 2013; 5: 36. 2013/08/21. https://doi.org/10.1186/alzrt195
Joseph-Mathurin N, Su Y, Blazey TM, Jasielec M, Vlassenko A, Friedrichsen K, Gordon BA, Hornbeck RC, Cash L, Ances BM, Veale T, Cash DM, Brickman AM, Buckles V, Cairns NJ, Cruchaga C, Goate A, Jack CR Jr, Karch C, Klunk W, Koeppe RA, Marcus DS, Mayeux R, McDade E, Noble JM, Ringman J, Saykin AJ, Thompson PM, Xiong C, Morris JC, Bateman RJ, Benzinger TLS, Dominantly Inherited Alzheimer Network (2018) Utility of perfusion PET measures to assess neuronal injury in Alzheimer’s disease. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring 10:669–677
Hyman BT, Phelps CH, Beach TG, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimer’s & dementia : the journal of the Alzheimer's Association 2012; 8: 1–13. 2012/01/24. https://doi.org/10.1016/j.jalz.2011.10.007
Nelson PT, Alafuzoff I, Bigio EH, Bouras C, Braak H, Cairns NJ, Castellani RJ, Crain BJ, Davies P, Tredici KD, Duyckaerts C, Frosch MP, Haroutunian V, Hof PR, Hulette CM, Hyman BT, Iwatsubo T, Jellinger KA, Jicha GA, Kövari E, Kukull WA, Leverenz JB, Love S, Mackenzie IR, Mann DM, Masliah E, McKee AC, Montine TJ, Morris JC, Schneider JA, Sonnen JA, Thal DR, Trojanowski JQ, Troncoso JC, Wisniewski T, Woltjer RL, Beach TG (2012) Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J Neuropathol Exp Neurol 71:362–381
Arriagada PV, Growdon JH, Hedley-Whyte ET, Hyman BT (1992) Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer’s disease. Neurology 42:631–631
Chien DT, Bahri S, Szardenings AK, et al. Early clinical PET imaging results with the novel PHF-tau radioligand [F-18]-T807. Journal of Alzheimer’s disease : JAD 2013; 34: 457–468. 2012/12/14. https://doi.org/10.3233/jad-122059
Johnson KA, Schultz A, Betensky RA, et al. Tau positron emission tomographic imaging in aging and early Alzheimer disease. Annals of neurology 2016; 79: 110–119. 2015/10/28. https://doi.org/10.1002/ana.24546
Xia CF, Arteaga J, Chen G, et al. [(18)F]T807, a novel tau positron emission tomography imaging agent for Alzheimer’s disease. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 2013; 9: 666–676. 2013/02/16. https://doi.org/10.1016/j.jalz.2012.11.008
Yaqub M, Tolboom N, Boellaard R, et al. Simplified parametric methods for [11C]PIB studies. NeuroImage 2008; 42: 76–86. 2008/06/11. https://doi.org/10.1016/j.neuroimage.2008.04.251
Rodriguez-Vieitez E, Leuzy A, Chiotis K, et al. Comparability of [(18)F]THK5317 and [(11)C]PIB blood flow proxy images with [(18)F]FDG positron emission tomography in Alzheimer’s disease. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism 2017; 37: 740–749. 2016/04/24. https://doi.org/10.1177/0271678x16645593
Peretti DE, Vállez García D, Reesink FE, et al. Diagnostic performance of regional cerebral blood flow images derived from dynamic PIB scans in Alzheimer’s disease. EJNMMI research 2019; 9: 59. 2019/07/06. https://doi.org/10.1186/s13550-019-0528-3
Chen YJ, Rosario BL, Mowrey W, et al. Relative 11C-PiB delivery as a proxy of relative CBF: quantitative evaluation using single-session 15O-water and 11C-PiB PET. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 2015; 56: 1199–1205. 2015/06/06. https://doi.org/10.2967/jnumed.114.152405
Ottoy J, Verhaeghe J, Niemantsverdriet E, et al. (18)F-FDG PET, the early phases and the delivery rate of (18)F-AV45 PET as proxies of cerebral blood flow in Alzheimer’s disease: validation against (15)O-H(2)O PET. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 2019; 15: 1172–1182. 2019/08/14. https://doi.org/10.1016/j.jalz.2019.05.010
Peretti DE, Vállez García D, Reesink FE, et al. Correction: Relative cerebral flow from dynamic PIB scans as an alternative for FDG scans in Alzheimer’s disease PET studies. PloS one 2019; 14: e0214187. 2019/03/19. https://doi.org/10.1371/journal.pone.0214187
van Berckel BN, Ossenkoppele R, Tolboom N, et al. Longitudinal amyloid imaging using 11C-PiB: methodologic considerations. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 2013; 54: 1570–1576. 2013/08/14. https://doi.org/10.2967/jnumed.112.113654
Golla SS, Timmers T, Ossenkoppele R et al (2017) Quantification of tau load using [18 F] AV1451 PET. Mol Imaging Biol 19:963–971
Golla SS, Wolters EE, Timmers T, et al. Parametric methods for [18F] flortaucipir PET. Journal of Cerebral Blood Flow & Metabolism 2018: 0271678X18820765
Timmers T, Ossenkoppele R, Visser D et al (2019) Test-retest repeatability of [18F]Flortaucipir PET in Alzheimer’s disease and cognitively normal individuals. J Cereb Blood Flow Metab in press in press
Wolters EE, Golla SS, Timmers T et al (2018) A novel partial volume correction method for accurate quantification of [18 F] flortaucipir in the hippocampus. EJNMMI Res 8:79
Timmers T, Ossenkoppele R, Wolters EE, Verfaillie SCJ, Visser D, Golla SSV, Barkhof F, Scheltens P, Boellaard R, van der Flier WM, van Berckel BNM (2019) Associations between quantitative [18 F] flortaucipir tau PET and atrophy across the Alzheimer’s disease spectrum. Alzheimers Res Ther 11:60
Barret O, Alagille D, Sanabria S, et al. Kinetic modeling of the tau PET tracer (18)F-AV-1451 in human healthy volunteers and Alzheimer disease subjects. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 2017; 58: 1124–1131. 2016/12/03. https://doi.org/10.2967/jnumed.116.182881
Boellaard R, van Lingen A, van Balen SC, et al. Characteristics of a new fully programmable blood sampling device for monitoring blood radioactivity during PET. European journal of nuclear medicine 2001; 28: 81–89. 2001/02/24. https://doi.org/10.1007/s002590000405
Hammers A, Allom R, Koepp MJ, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Human brain mapping 2003; 19: 224–247. 2003/07/23. https://doi.org/10.1002/hbm.10123
Svarer C, Madsen K, Hasselbalch SG, et al. MR-based automatic delineation of volumes of interest in human brain PET images using probability maps. NeuroImage 2005; 24: 969–979. 2005/01/27. https://doi.org/10.1016/j.neuroimage.2004.10.017
Lammertsma AA and Hume SP. Simplified reference tissue model for PET receptor studies. NeuroImage 1996; 4: 153–158. 1996/12/01. https://doi.org/10.1006/nimg.1996.0066
Gunn RN, Lammertsma AA, Hume SP, et al. Parametric imaging of ligand-receptor binding in PET using a simplified reference region model. NeuroImage 1997; 6: 279–287. 1998/02/07. https://doi.org/10.1006/nimg.1997.0303
Golla SSV, Timmers T, Ossenkoppele R, et al. Quantification of tau load using [(18)F]AV1451 PET. Molecular imaging and biology 2017; 19: 963–971. 2017/04/05. https://doi.org/10.1007/s11307-017-1080-z
Heeman F, Yaqub M, Lopes Alves I, et al. Optimized dual-time-window protocols for quantitative [(18)F]flutemetamol and [(18)F]florbetaben PET studies. EJNMMI research 2019; 9: 32. 2019/03/29. https://doi.org/10.1186/s13550-019-0499-4
Hammes J, Leuwer I, Bischof GN, et al. Multimodal correlation of dynamic [(18)F]-AV-1451 perfusion PET and neuronal hypometabolism in [(18)F]-FDG PET. European journal of nuclear medicine and molecular imaging 2017; 44: 2249–2256. 2017/10/14. https://doi.org/10.1007/s00259-017-3840-z
Acknowledgements
Research of Amsterdam Alzheimer Center is part of the Neurodegeneration program of Amsterdam Neuroscience. [18F]Flortaucipir PET scans were sponsored by Avid Radiopharmaceuticals Inc.
Author information
Authors and Affiliations
Contributions
HT, DV, SG, MY, BB and RB contributed to the concept and design of the study. EW, TT and DV contributed to the acquisition of the data. HT, SG, MY and RB contributed to the analysis and interpretation of the data. HT drafted the manuscript. HT, SG, EW, TT, WF, RO, MY, BB and RB read, critically reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tuncel, H., Visser, D., Yaqub, M. et al. Effect of Shortening the Scan Duration on Quantitative Accuracy of [18F]Flortaucipir Studies. Mol Imaging Biol 23, 604–613 (2021). https://doi.org/10.1007/s11307-021-01581-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-021-01581-5