Abstract
Background
Metabolomics provides measurement of numerous metabolites in human samples, which can be a useful tool in clinical research. Blood and urine are regarded as preferred subjects of study because of their minimally invasive collection and simple preprocessing methods. Adhering to standard operating procedures is an essential factor in ensuring excellent sample quality and reliable results.
Aim of review
In this review, we summarize the studies about the impacts of various preprocessing factors on metabolomics studies involving clinical blood and urine samples in order to provide guidance for sample collection and preprocessing.
Key scientific concepts of review
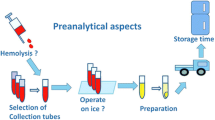
Clinical information is important for sample grouping and data analysis which deserves attention before sample collection. Plasma and serum as well as urine samples are appropriate for metabolomics analysis. Collection tubes, hemolysis, delay at room temperature, and freeze–thaw cycles may affect metabolic profiles of blood samples. Collection time, time between sampling and examination, contamination, normalization strategies, and storage conditions may alter analysis results of urine samples. Taking these collection and preprocessing factors into account, this review provides suggestions of standard sample preprocessing.
Similar content being viewed by others
References
Ackermann, D., Groessl, M., Pruijm, M., Ponte, B., Escher, G., d'Uscio, C. H., et al. (2019). Reference intervals for the urinary steroid metabolome: The impact of sex, age, day and night time on human adult steroidogenesis. PLoS ONE,14(3), e0214549. https://doi.org/10.1371/journal.pone.0214549.
Alberice, J. V., Amaral, A. F., Armitage, E. G., Lorente, J. A., Algaba, F., Carrilho, E., et al. (2013). Searching for urine biomarkers of bladder cancer recurrence using a liquid chromatography-mass spectrometry and capillary electrophoresis-mass spectrometry metabolomics approach. Journal of Chromatography A,1318, 163–170. https://doi.org/10.1016/j.chroma.2013.10.002.
Amberg, A., Riefke, B., Schlotterbeck, G., Ross, A., Senn, H., Dieterle, F., et al. (2017). NMR and MS methods for metabolomics. Methods in Molecular Biology,1641, 229–258. https://doi.org/10.1007/978-1-4939-7172-5_13.
Ammerlaan, W., Trezzi, J. P., Lescuyer, P., Mathay, C., Hiller, K., & Betsou, F. (2014). Method validation for preparing serum and plasma samples from human blood for downstream proteomic, metabolomic, and circulating nucleic acid-based applications. Biopreserv Biobank,12(4), 269–280. https://doi.org/10.1089/bio.2014.0003.
Anton, G., Wilson, R., Yu, Z. H., Prehn, C., Zukunft, S., Adamski, J., et al. (2015). Pre-analytical sample quality: Metabolite ratios as an intrinsic marker for prolonged room temperature exposure of serum samples. PLoS ONE,10(3), e0121495. https://doi.org/10.1371/journal.pone.0121495.
Bando, K., Kawahara, R., Kunimatsu, T., Sakai, J., Kimura, J., Funabashi, H., et al. (2010). Influences of biofluid sample collection and handling procedures on GC-MS based metabolomic studies. Journal of Bioscience and Bioengineering,110(4), 491–499. https://doi.org/10.1016/j.jbiosc.2010.04.010.
Barri, T., & Dragsted, L. O. (2013). UPLC-ESI-QTOF/MS and multivariate data analysis for blood plasma and serum metabolomics: Effect of experimental artefacts and anticoagulant. Analytica Chimica Acta,768, 118–128. https://doi.org/10.1016/j.aca.2013.01.015.
Barton, R. H., Nicholson, J. K., Elliott, P., & Holmes, E. (2008). High-throughput 1H NMR-based metabolic analysis of human serum and urine for large-scale epidemiological studies: Validation study. International Journal of Epidemiology,37(Suppl 1), i31–40. https://doi.org/10.1093/ije/dym284.
Bernini, P., Bertini, I., Luchinat, C., Nincheri, P., Staderini, S., & Turano, P. (2011). Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. Journal of Biomolecular NMR,49(3–4), 231–243. https://doi.org/10.1007/s10858-011-9489-1.
Bervoets, L., Louis, E., Reekmans, G., Mesotten, L., Thomeer, M., Adriaensens, P., et al. (2015). Influence of preanalytical sampling conditions on the 1H NMR metabolic profile of human blood plasma and introduction of the Standard PREanalytical Code used in biobanking. Metabolomics,11(5), 1197–1207. https://doi.org/10.1007/s11306-015-0774-y.
Bouatra, S., Aziat, F., Mandal, R., Guo, A. C., Wilson, M. R., Knox, C., et al. (2013). The human urine metabolome. PLoS ONE,8(9), e73076. https://doi.org/10.1371/journal.pone.0073076.
Breier, M., Wahl, S., Prehn, C., Fugmann, M., Ferrari, U., Weise, M., et al. (2014). Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS ONE,9(2), e89728. https://doi.org/10.1371/journal.pone.0089728.
Brunius, C., Pedersen, A., Malmodin, D., Karlsson, B. G., Andersson, L. I., Tybring, G., et al. (2017). Prediction and modeling of pre-analytical sampling errors as a strategy to improve plasma NMR metabolomics data. Bioinformatics,33(22), 3567–3574. https://doi.org/10.1093/bioinformatics/btx442.
Budde, K., Gok, O. N., Pietzner, M., Meisinger, C., Leitzmann, M., Nauck, M., et al. (2016). Quality assurance in the pre-analytical phase of human urine samples by (1)H NMR spectroscopy. Archives of Biochemistry and Biophysics,589, 10–17. https://doi.org/10.1016/j.abb.2015.07.016.
Chadha, V., Garg, U., & Alon, U. S. (2001). Measurement of urinary concentration: A critical appraisal of methodologies. Pediatric Nephrology (Berlin, Germany),16(4), 374–382. https://doi.org/10.1007/s004670000551.
Chang, C. J., Yang, J. Y., Xia, W., Chen, C. T., Xie, X., Chao, C. H., et al. (2011). EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell,19(1), 86–100. https://doi.org/10.1016/j.ccr.2010.10.035.
Chetwynd, A. J., Abdul-Sada, A., Holt, S. G., & Hill, E. M. (2016). Use of a pre-analysis osmolality normalisation method to correct for variable urine concentrations and for improved metabolomic analyses. Journal of Chromatography A,1431, 103–110. https://doi.org/10.1016/j.chroma.2015.12.056.
Comstock, G. W., Burke, A. E., Norkus, E. P., Gordon, G. B., Hoffman, S. C., & Helzlsouer, K. J. (2008). Effects of repeated freeze-thaw cycles on concentrations of cholesterol, micronutrients, and hormones in human plasma and serum. American Journal of Epidemiology,168(7), 827–830. https://doi.org/10.1093/aje/kwn327.
Cuhadar, S., Atay, A., Koseoglu, M., Dirican, A., & Hur, A. (2012). Stability studies of common biochemical analytes in serum separator tubes with or without gel barrier subjected to various storage conditions. Biochemia Medica (Zagreb),22(2), 202–214. https://doi.org/10.11613/bm.2012.023.
de Sousa, E. B., Dos Santos, G. C. J., Duarte, M. E. L., Moura, V. N., & Aguiar, D. P. (2017). Metabolomics as a promising tool for early osteoarthritis diagnosis. Brazilian Journal of Medical and Biological Research,50(11), e6485. https://doi.org/10.1590/1414-431X20176485.
Delanghe, J. R., & Speeckaert, M. M. (2016). Preanalytics in urinalysis. Clinical Biochemistry,49(18), 1346–1350. https://doi.org/10.1016/j.clinbiochem.2016.10.016.
Denery, J. R., Nunes, A. A., & Dickerson, T. J. (2011). Characterization of differences between blood sample matrices in untargeted metabolomics. Analytical Chemistry,83(3), 1040–1047. https://doi.org/10.1021/ac102806p.
Denihan, N. M., Walsh, B. H., Reinke, S. N., Sykes, B. D., Mandal, R., Wishart, D. S., et al. (2015). The effect of haemolysis on the metabolomic profile of umbilical cord blood. Clinical Biochemistry,48(7–8), 534–537. https://doi.org/10.1016/j.clinbiochem.2015.02.004.
Deprez, S., Sweatman, B. C., Connor, S. C., Haselden, J. N., & Waterfield, C. J. (2002). Optimisation of collection, storage and preparation of rat plasma for 1H NMR spectroscopic analysis in toxicology studies to determine inherent variation in biochemical profiles. Journal of Pharmaceutical and Biomedical Analysis,30(4), 1297–1310. https://doi.org/10.1016/s0731-7085(02)00455-7.
Dettmer, K., Almstetter, M. F., Appel, I. J., Nurnberger, N., Schlamberger, G., Gronwald, W., et al. (2010). Comparison of serum versus plasma collection in gas chromatography–mass spectrometry-based metabolomics. Electrophoresis,31(14), 2365–2373. https://doi.org/10.1002/elps.200900778.
Di Gregorio, M. C., Jager, A. V., Costa, A. A., Bordin, K., Rottinhghaus, G. E., Petta, T., et al. (2017). Determination of aflatoxin B1-lysine in pig serum and plasma by liquid chromatography-tandem mass spectrometry. Journal of Analytical Toxicology,41(3), 236–241. https://doi.org/10.1093/jat/bkw126.
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols,6(7), 1060–1083. https://doi.org/10.1038/nprot.2011.335.
Dunn, W. B., Broadhurst, D., Ellis, D. I., Brown, M., Halsall, A., O'Hagan, S., et al. (2008). A GC-TOF-MS study of the stability of serum and urine metabolomes during the UK Biobank sample collection and preparation protocols. International Journal of Epidemiology,37(Suppl 1), i23–30. https://doi.org/10.1093/ije/dym281.
Ellervik, C., & Vaught, J. (2015). Preanalytical variables affecting the integrity of human biospecimens in biobanking. Clinical Chemistry,61(7), 914–934. https://doi.org/10.1373/clinchem.2014.228783.
Emwas, A. H., Roy, R., McKay, R. T., Ryan, D., Brennan, L., Tenori, L., et al. (2016). Recommendations and standardization of biomarker quantification using NMR-based metabolomics with particular focus on urinary analysis. Journal of Proteome Research,15(2), 360–373. https://doi.org/10.1021/acs.jproteome.5b00885.
Fan, Y., Zhou, X., Xia, T. S., Chen, Z., Li, J., Liu, Q., et al. (2016). Human plasma metabolomics for identifying differential metabolites and predicting molecular subtypes of breast cancer. Oncotarget,7(9), 9925–9938. https://doi.org/10.18632/oncotarget.7155.
Fernández-Peralbo, M. A., & Luque de Castro, M. D. (2012). Preparation of urine samples prior to targeted or untargeted metabolomics mass-spectrometry analysis. TrAC Trends in Analytical Chemistry,41, 75–85. https://doi.org/10.1016/j.trac.2012.08.011.
Filzmoser, P., & Walczak, B. (2014). What can go wrong at the data normalization step for identification of biomarkers? Journal of Chromatography A,1362, 194–205. https://doi.org/10.1016/j.chroma.2014.08.050.
Fliniaux, O., Gaillard, G., Lion, A., Cailleu, D., Mesnard, F., & Betsou, F. (2011). Influence of common preanalytical variations on the metabolic profile of serum samples in biobanks. Journal of Biomolecular NMR,51(4), 457–465. https://doi.org/10.1007/s10858-011-9574-5.
Fobker, M. (2014). Stability of glucose in plasma with different anticoagulants. Clinical Chemistry and Laboratory Medicine,52(7), 1057–1060. https://doi.org/10.1515/cclm-2013-1049.
Gagnebin, Y., Tonoli, D., Lescuyer, P., Ponte, B., de Seigneux, S., Martin, P. Y., et al. (2017). Metabolomic analysis of urine samples by UHPLC-QTOF-MS: Impact of normalization strategies. Analytica Chimica Acta,955, 27–35. https://doi.org/10.1016/j.aca.2016.12.029.
Garcia-Villalba, R., Carrasco-Pancorbo, A., Nevedomskaya, E., Mayboroda, O. A., Deelder, A. M., Segura-Carretero, A., et al. (2010). Exploratory analysis of human urine by LC-ESI-TOF MS after high intake of olive oil: Understanding the metabolism of polyphenols. Analytical and Bioanalytical Chemistry,398(1), 463–475. https://doi.org/10.1007/s00216-010-3899-x.
Gibney, M. J., Walsh, M., Brennan, L., Roche, H. M., German, B., & van Ommen, B. (2005). Metabolomics in human nutrition: Opportunities and challenges. American Journal of Clinical Nutrition,82(3), 497–503. https://doi.org/10.1093/ajcn.82.3.497.
Gika, H. G., Theodoridis, G. A., & Wilson, I. D. (2008). Liquid chromatography and ultra-performance liquid chromatography-mass spectrometry fingerprinting of human urine: Sample stability under different handling and storage conditions for metabonomics studies. Journal of Chromatography A,1189(1–2), 314–322. https://doi.org/10.1016/j.chroma.2007.10.066.
Gika, H. G., Theodoridis, G. A., Wingate, J. E., & Wilson, I. D. (2007). Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: Application to human urine. Journal of Proteome Research,6(8), 3291–3303. https://doi.org/10.1021/pr070183p.
Giskeodegard, G. F., Davies, S. K., Revell, V. L., Keun, H., & Skene, D. J. (2015). Diurnal rhythms in the human urine metabolome during sleep and total sleep deprivation. Scientific Reports,5, 14843. https://doi.org/10.1038/srep14843.
Gonzalez-Covarrubias, V., Dane, A., Hankemeier, T., & Vreeken, R. J. (2013). The influence of citrate, EDTA, and heparin anticoagulants to human plasma LC-MS lipidomic profiling. Metabolomics,9(2), 337–348. https://doi.org/10.1007/s11306-012-0450-4.
Griffin, J. L., & Bollard, M. E. (2004). Metabonomics: its potential as a tool in toxicology for safety assessment and data integration. Current Drug Metabolism,5(5), 389–398. https://doi.org/10.2174/1389200043335432.
Haid, M., Muschet, C., Wahl, S., Romisch-Margl, W., Prehn, C., Moller, G., et al. (2018). Long-term stability of human plasma metabolites during storage at -80 degrees C. Journal of Proteome Research,17(1), 203–211. https://doi.org/10.1021/acs.jproteome.7b00518.
Hebels, D. G., Georgiadis, P., Keun, H. C., Athersuch, T. J., Vineis, P., Vermeulen, R., et al. (2013). Performance in omics analyses of blood samples in long-term storage: Opportunities for the exploitation of existing biobanks in environmental health research. Environmental Health Perspectives,121(4), 480–487. https://doi.org/10.1289/ehp.1205657.
Hernandes, V. V., Barbas, C., & Dudzik, D. (2017). A review of blood sample handling and pre-processing for metabolomics studies. Electrophoresis,38(18), 2232–2241. https://doi.org/10.1002/elps.201700086.
Hirayama, A., Sugimoto, M., Suzuki, A., Hatakeyama, Y., Enomoto, A., Harada, S., et al. (2015). Effects of processing and storage conditions on charged metabolomic profiles in blood. Electrophoresis,36(18), 2148–2155. https://doi.org/10.1002/elps.201400600.
Ishikawa, M., Maekawa, K., Saito, K., Senoo, Y., Urata, M., Murayama, M., et al. (2014). Plasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects' gender and age. PLoS ONE,9(3), e91806. https://doi.org/10.1371/journal.pone.0091806.
Jain, M., Kennedy, A. D., Elsea, S. H., & Miller, M. J. (2017). Analytes related to erythrocyte metabolism are reliable biomarkers for preanalytical error due to delayed plasma processing in metabolomics studies. Clinica Chimica Acta,466, 105–111. https://doi.org/10.1016/j.cca.2017.01.005.
Jobard, E., Tredan, O., Postoly, D., Andre, F., Martin, A. L., Elena-Herrmann, B., et al. (2016). A systematic evaluation of blood serum and plasma pre-analytics for metabolomics cohort studies. International Journal of Molecular Sciences,17(12), 2035. https://doi.org/10.3390/ijms17122035.
Kaluarachchi, M., Boulange, C. L., Karaman, I., Lindon, J. C., Ebbels, T. M. D., Elliott, P., et al. (2018). A comparison of human serum and plasma metabolites using untargeted (1)H NMR spectroscopy and UPLC-MS. Metabolomics,14(3), 32. https://doi.org/10.1007/s11306-018-1332-1.
Kamlage, B., Maldonado, S. G., Bethan, B., Peter, E., Schmitz, O., Liebenberg, V., et al. (2014). Quality markers addressing preanalytical variations of blood and plasma processing identified by broad and targeted metabolite profiling. Clinical Chemistry,60(2), 399–412. https://doi.org/10.1373/clinchem.2013.211979.
Keller, B. O., Sui, J., Young, A. B., & Whittal, R. M. (2008). Interferences and contaminants encountered in modern mass spectrometry. Analytica Chimica Acta,627(1), 71–81. https://doi.org/10.1016/j.aca.2008.04.043.
Kim, K., Mall, C., Taylor, S. L., Hitchcock, S., Zhang, C., Wettersten, H. I., et al. (2014). Mealtime, temporal, and daily variability of the human urinary and plasma metabolomes in a tightly controlled environment. PLoS ONE,9(1), e86223. https://doi.org/10.1371/journal.pone.0086223.
Kirwan, J. A., Brennan, L., Broadhurst, D., Fiehn, O., Cascante, M., Dunn, W. B., et al. (2018). Preanalytical processing and biobanking procedures of biological samples for metabolomics research: A white paper, community perspective (for "precision medicine and pharmacometabolomics task group"-the metabolomics society initiative). Clinical Chemistry,64(8), 1158–1182. https://doi.org/10.1373/clinchem.2018.287045.
La Frano, M. R., Carmichael, S. L., Ma, C., Hardley, M., Shen, T., Wong, R., et al. (2018). Impact of post-collection freezing delay on the reliability of serum metabolomics in samples reflecting the California mid-term pregnancy biobank. Metabolomics,14(11), 151. https://doi.org/10.1007/s11306-018-1450-9.
Lauridsen, M., Hansen, S. H., Jaroszewski, J. W., & Cornett, C. (2007). Human urine as test material in 1H NMR-based metabonomics: Recommendations for sample preparation and storage. Analytical Chemistry,79(3), 1181–1186. https://doi.org/10.1021/ac061354x.
Lawton, K. A., Berger, A., Mitchell, M., Milgram, K. E., Evans, A. M., Guo, L., et al. (2008). Analysis of the adult human plasma metabolome. Pharmacogenomics,9(4), 383–397. https://doi.org/10.2217/14622416.9.4.383.
Lee, J. E., & Kim, Y. Y. (2017). Impact of preanalytical variations in blood-derived biospecimens on omics studies: Toward precision biobanking? OMICS: A Journal of Integrative Biology,21(9), 499–508. https://doi.org/10.1089/omi.2017.0109.
Lehmann, R. (2015). Preanalytics: What can metabolomics learn from clinical chemistry? Bioanalysis,7(8), 927–930. https://doi.org/10.4155/bio.15.23.
Lesche, D., Geyer, R., Lienhard, D., Nakas, C. T., Diserens, G., Vermathen, P., et al. (2016). Does centrifugation matter? Centrifugal force and spinning time alter the plasma metabolome. Metabolomics,12(10), 159. https://doi.org/10.1007/s11306-016-1109-3.
Li, N., Song, Y., Tang, H., & Wang, Y. (2016). Recent developments in sample preparation and data pre-treatment in metabonomics research. Archives of Biochemistry and Biophysics,589, 4–9. https://doi.org/10.1016/j.abb.2015.08.024.
Lippi, G., Blanckaert, N., Bonini, P., Green, S., Kitchen, S., Palicka, V., et al. (2008). Haemolysis: An overview of the leading cause of unsuitable specimens in clinical laboratories. Clinical Chemistry and Laboratory Medicine,46(6), 764–772. https://doi.org/10.1515/CCLM.2008.170.
Liu, X., Hoene, M., Wang, X., Yin, P., Haring, H. U., Xu, G., et al. (2018). Serum or plasma, what is the difference? Investigations to facilitate the sample material selection decision making process for metabolomics studies and beyond. Analytica Chimica Acta,1037, 293–300. https://doi.org/10.1016/j.aca.2018.03.009.
Lopez-Bascon, M. A., Priego-Capote, F., Peralbo-Molina, A., Calderon-Santiago, M., & Luque de Castro, M. D. (2016). Influence of the collection tube on metabolomic changes in serum and plasma. Talanta,150, 681–689. https://doi.org/10.1016/j.talanta.2015.12.079.
Malm, L., Tybring, G., Moritz, T., Landin, B., & Galli, J. (2016). Metabolomic quality assessment of EDTA plasma and serum samples. Biopreserv Biobank,14(5), 416–423. https://doi.org/10.1089/bio.2015.0092.
Mei, H., Hsieh, Y., Nardo, C., Xu, X., Wang, S., Ng, K., et al. (2003). Investigation of matrix effects in bioanalytical high-performance liquid chromatography/tandem mass spectrometric assays: Application to drug discovery. Rapid Communications in Mass Spectrometry,17(1), 97–103. https://doi.org/10.1002/rcm.876.
Miller, R. C., Brindle, E., Holman, D. J., Shofer, J., Klein, N. A., Soules, M. R., et al. (2004). Comparison of specific gravity and creatinine for normalizing urinary reproductive hormone concentrations. Clinical Chemistry,50(5), 924–932. https://doi.org/10.1373/clinchem.2004.032292.
Minami, Y., Kasukawa, T., Kakazu, Y., Iigo, M., Sugimoto, M., Ikeda, S., et al. (2009). Measurement of internal body time by blood metabolomics. Proceedings of the National academy of Sciences of the United States of America,106(24), 9890–9895. https://doi.org/10.1073/pnas.0900617106.
Morris, C., O'Grada, C., Ryan, M., Roche, H. M., Gibney, M. J., Gibney, E. R., et al. (2012). The relationship between BMI and metabolomic profiles: A focus on amino acids. The Proceedings of the Nutrition Society,71(4), 634–638. https://doi.org/10.1017/S0029665112000699.
Nicholson, J. K., Buckingham, M. J., & Sadler, P. J. (1983). High resolution 1H n.m.r. studies of vertebrate blood and plasma. Biochemical Journal,211(3), 605–615. https://doi.org/10.1042/bj2110605.
Nishiumi, S., Suzuki, M., Kobayashi, T., & Yoshida, M. (2018). Differences in metabolite profiles caused by pre-analytical blood processing procedures. Journal of Bioscience and Bioengineering,125(5), 613–618. https://doi.org/10.1016/j.jbiosc.2017.11.011.
Paglia, G., Del Greco, F. M., Sigurdsson, B. B., Rainer, J., Volani, C., Hicks, A. A., et al. (2018). Influence of collection tubes during quantitative targeted metabolomics studies in human blood samples. Clinica Chimica Acta,486, 320–328. https://doi.org/10.1016/j.cca.2018.08.014.
Palmas, F., Mussap, M., & Fattuoni, C. (2018). Urine metabolome analysis by gas chromatography-mass spectrometry (GC-MS): Standardization and optimization of protocols for urea removal and short-term sample storage. Clinica Chimica Acta,485, 236–242. https://doi.org/10.1016/j.cca.2018.07.006.
Pinto, J., Domingues, M. R., Galhano, E., Pita, C., Almeida Mdo, C., Carreira, I. M., et al. (2014). Human plasma stability during handling and storage: Impact on NMR metabolomics. Analyst,139(5), 1168–1177. https://doi.org/10.1039/c3an02188b.
Pouralijan Amiri, M., Khoshkam, M., Salek, R. M., Madadi, R., Faghanzadeh Ganji, G., & Ramazani, A. (2019). Metabolomics in early detection and prognosis of acute coronary syndrome. Clinica Chimica Acta,495, 43–53. https://doi.org/10.1016/j.cca.2019.03.1632.
Psychogios, N., Hau, D. D., Peng, J., Guo, A. C., Mandal, R., Bouatra, S., et al. (2011). The human serum metabolome. PLoS ONE,6(2), e16957. https://doi.org/10.1371/journal.pone.0016957.
Rotter, M., Brandmaier, S., Prehn, C., Adam, J., Rabstein, S., Gawrych, K., et al. (2017). Stability of targeted metabolite profiles of urine samples under different storage conditions. Metabolomics,13(1), 4. https://doi.org/10.1007/s11306-016-1137-z.
Salvagno, G. L., Danese, E., & Lippi, G. (2017). Preanalytical variables for liquid chromatography-mass spectrometry (LC-MS) analysis of human blood specimens. Clinical Biochemistry,50(10–11), 582–586. https://doi.org/10.1016/j.clinbiochem.2017.04.012.
Saude, E. J., & Sykes, B. D. (2007). Urine stability for metabolomic studies: Effects of preparation and storage. Metabolomics,3(1), 19–27. https://doi.org/10.1007/s11306-006-0042-2.
Scalbert, A., Brennan, L., Fiehn, O., Hankemeier, T., Kristal, B. S., van Ommen, B., et al. (2009). Mass-spectrometry-based metabolomics: Limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics,5(4), 435–458. https://doi.org/10.1007/s11306-009-0168-0.
Silvester, S., & Zang, F. (2012). Overcoming non-specific adsorption issues for AZD9164 in human urine samples: Consideration of bioanalytical and metabolite identification procedures. Journal of Chromatography B,893–894, 134–143. https://doi.org/10.1016/j.jchromb.2012.03.004.
Siskos, A. P., Jain, P., Romisch-Margl, W., Bennett, M., Achaintre, D., Asad, Y., et al. (2017). Interlaboratory reproducibility of a targeted metabolomics platform for analysis of human serum and plasma. Analytical Chemistry,89(1), 656–665. https://doi.org/10.1021/acs.analchem.6b02930.
Soldi, L. R., Maltos, A. L., da Cunha, D. F., & Portari, G. V. (2018). Correlation between first morning single void and 24-hour urines: The reliability to quantify niacin status. Medical Science Monitor Basic Research,24, 206–209. https://doi.org/10.12659/MSMBR.910087.
Teahan, O., Gamble, S., Holmes, E., Waxman, J., Nicholson, J. K., Bevan, C., et al. (2006). Impact of analytical bias in metabonomic studies of human blood serum and plasma. Analytical Chemistry,78(13), 4307–4318. https://doi.org/10.1021/ac051972y.
Townsend, M. K., Bao, Y., Poole, E. M., Bertrand, K. A., Kraft, P., Wolpin, B. M., et al. (2016). Impact of pre-analytic blood sample collection factors on metabolomics. Cancer Epidemiology, Biomarkers & Prevention,25(5), 823–829. https://doi.org/10.1158/1055-9965.EPI-15-1206.
van der Sar, S. A., Zielman, R., Terwindt, G. M., van den Maagdenberg, A. M., Deelder, A. M., Mayboroda, O. A., et al. (2015). Ethanol contamination of cerebrospinal fluid during standardized sampling and its effect on (1)H-NMR metabolomics. Analytical and Bioanalytical Chemistry,407(16), 4835–4839. https://doi.org/10.1007/s00216-015-8663-9.
Waikar, S. S., Sabbisetti, V. S., & Bonventre, J. V. (2010). Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate. Kidney International,78(5), 486–494. https://doi.org/10.1038/ki.2010.165.
Wang, Y., Carter, B. D., Gapstur, S. M., McCullough, M. L., Gaudet, M. M., & Stevens, V. L. (2018). Reproducibility of non-fasting plasma metabolomics measurements across processing delays. Metabolomics,14(10), 129. https://doi.org/10.1007/s11306-018-1429-6.
Warrack, B. M., Hnatyshyn, S., Ott, K. H., Reily, M. D., Sanders, M., Zhang, H., et al. (2009). Normalization strategies for metabonomic analysis of urine samples. Journal of Chromatography B,877(5–6), 547–552. https://doi.org/10.1016/j.jchromb.2009.01.007.
Weigert, C., Lehmann, R., Hartwig, S., & Lehr, S. (2014). The secretome of the working human skeletal muscle—A promising opportunity to combat the metabolic disaster? Proteomics Clinical Applications,8(1–2), 5–18. https://doi.org/10.1002/prca.201300094.
Wen, C. P., Zhang, F., Liang, D., Wen, C., Gu, J., Skinner, H., et al. (2015). The ability of bilirubin in identifying smokers with higher risk of lung cancer: A large cohort study in conjunction with global metabolomic profiling. Clinical Cancer Research,21(1), 193–200. https://doi.org/10.1158/1078-0432.CCR-14-0748.
Wojakowska, A., Chekan, M., Marczak, L., Polanski, K., Lange, D., Pietrowska, M., et al. (2015). Detection of metabolites discriminating subtypes of thyroid cancer: Molecular profiling of FFPE samples using the GC/MS approach. Molecular and Cellular Endocrinology,417, 149–157. https://doi.org/10.1016/j.mce.2015.09.021.
Yao, C. H., Liu, G. Y., Yang, K., Gross, R. W., & Patti, G. J. (2016). Inaccurate quantitation of palmitate in metabolomics and isotope tracer studies due to plastics. Metabolomics. https://doi.org/10.1007/s11306-016-1081-y.
Yin, P., Lehmann, R., & Xu, G. (2015). Effects of pre-analytical processes on blood samples used in metabolomics studies. Analytical and Bioanalytical Chemistry,407(17), 4879–4892. https://doi.org/10.1007/s00216-015-8565-x.
Yin, P., Peter, A., Franken, H., Zhao, X., Neukamm, S. S., Rosenbaum, L., et al. (2013). Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clinical Chemistry,59(5), 833–845. https://doi.org/10.1373/clinchem.2012.199257.
Yu, Z., Kastenmuller, G., He, Y., Belcredi, P., Moller, G., Prehn, C., et al. (2011). Differences between human plasma and serum metabolite profiles. PLoS ONE,6(7), e21230. https://doi.org/10.1371/journal.pone.0021230.
Yuille, M., Illig, T., Hveem, K., Schmitz, G., Hansen, J., Neumaier, M., et al. (2010). Laboratory management of samples in biobanks: European consensus expert group report. Biopreserv Biobank,8(1), 65–69. https://doi.org/10.1089/bio.2010.8102.
Zamora-Ros, R., Rabassa, M., Cherubini, A., Urpi-Sarda, M., Llorach, R., Bandinelli, S., et al. (2011). Comparison of 24-h volume and creatinine-corrected total urinary polyphenol as a biomarker of total dietary polyphenols in the Invecchiare InCHIANTI study. Analytica Chimica Acta,704(1–2), 110–115. https://doi.org/10.1016/j.aca.2011.07.035.
Zhang, A., Sun, H., Wu, X., & Wang, X. (2012). Urine metabolomics. Clinica Chimica Acta,414, 65–69. https://doi.org/10.1016/j.cca.2012.08.016.
Zivkovic, A. M., Wiest, M. M., Nguyen, U. T., Davis, R., Watkins, S. M., & German, J. B. (2009). Effects of sample handling and storage on quantitative lipid analysis in human serum. Metabolomics,5(4), 507–516. https://doi.org/10.1007/s11306-009-0174-2.
Funding
This research was supported by NSFC (National Natural Science Fundation of China, Nos. 81972966, 81672091) and BJNSF (Natural Science Fundation of Beijing, No. 7172232).
Author information
Authors and Affiliations
Contributions
LM and LX conceived of the concept of the review. HB and XJ conducted the literature search. XJ and ZG analyzed the search results and wrote the paper. ZG, HB, HL and LX were involved in revision.
Corresponding authors
Ethics declarations
Conflict of interest:
The authors declare that they have no conflict of interest.
Ethical approval:
This article does not contain any studies with human and/or animal participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bi, H., Guo, Z., Jia, X. et al. The key points in the pre-analytical procedures of blood and urine samples in metabolomics studies. Metabolomics 16, 68 (2020). https://doi.org/10.1007/s11306-020-01666-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-020-01666-2