Abstract
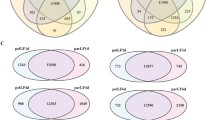
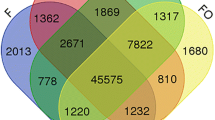
Hazel (Corylus spp.) has unique ovary and ovule development characteristics, as the ovary, ovule, and mature embryo sac only begin to develop after pollination. Frequent development failure of ovary and ovule causes yield losses in hazelnut. Despite the economic importance, proteins involved in the regulation of ovary and ovule differentiation remain largely unknown. To address these gaps, total protein was extracted from the pistillate inflorescences or young ovaries at three stages: stage F (unpollinated inflorescences), stage S (beginning of a rudimentary ovary formation), and stage T (ovule differentiation). Protein profiling analysis was carried out using isobaric tags for relative and absolute quantitation (iTRAQ), leading to the identification of 2586 proteins. In total, 815 differentially expressed proteins (DEPs) were identified, including 386, 173, and 697 DEPs in S-vs-F, T-vs-S, and T-vs-F paired comparisons, respectively. The proteins and DEPs using iTRAQ technology showed substantially higher transcript abundance than differentially expressed genes (DEGs) and unigenes in previous comparative transcriptome analysis at mRNA level. Pathway mapping results revealed that nine pathways were significantly enriched, sharing two common KEGG pathways, ko00940 (phenylpropanoid biosynthesis) and ko00941 (flavonoid biosynthesis), which were reported previously in a comparative transcriptome analysis. Of 815 DEPs, 236 were simultaneously differentially expressed at mRNA level, and a set of DEPs including chalcone synthase was predicted to participate in ovary and ovule differentiation based on the results at expression and mRNA level. Our findings provide new insight into the molecular mechanisms of ovary differentiation and development after pollination at the expression level.









Similar content being viewed by others
References
Aarts MG, Keijzer CJ, Stiekema WJ et al (1995) Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7:2115–2127. https://doi.org/10.1105/tpc.7.12.2115
Abdi N, Holford P, McGlasson B (2002) Application of two-dimensional gel electrophoresis to detect proteins associated with harvest maturity in stonefruit. Postharvest Biol Technol 26:1–13. https://doi.org/10.1016/S0925-5214(01)00197-1
Amaral JS, Casal S, Seabra RM, Oliveira BPP (2006) Effects of roasting on hazelnut lipids. J Agric Food Chem 54:1315–1321. https://doi.org/10.1021/jf052287v
Bagnoli F, Giannino D, Caparrini S et al (2002) Molecular cloning, characterisation and expression of a manganese superoxide dismutase gene from peach (Prunus persica [L.] Batsch). Mol Gen Genomics 267:321–328. https://doi.org/10.1007/s00438-002-0664-7
Beyhan N, Marangoz D (2007) An investigation of the relationship between reproductive growth and yield loss in hazelnut. Sci Hortic 113:208–215. https://doi.org/10.1016/j.scienta.2007.02.007
Bourdenx B, Bernard A, Domergue F, Pascal S, Leger A, Roby D, Pervent M, Vile D, Haslam RP, Napier JA, Lessire R, Joubes J (2011) Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol 156:29–45. https://doi.org/10.1104/pp.111.172320
Briat JF, Ravet K, Arnaud N, Duc C, Boucherez J, Touraine B, Cellier F, Gaymard F (2010) New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann Bot 105:811–822. https://doi.org/10.1093/aob/mcp128
Carter CJ, Thornburg RW (2004) Tobacco Nectarin III is a bifunctional enzyme with monodehydroascorbate reductase and carbonic anhydrase activities. Plant Mol Biol 54:415–425. https://doi.org/10.1023/B:PLAN.0000036373.84579.13
Chen JH, Jiang HW, Hsieh EJ, Chen HY, Chien CT, Hsieh HL, Lin TP (2012) Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol 158:340–351. https://doi.org/10.1104/pp.111.181875
Cheng Y, Zhang Y, Liu C, Ai P, Liu J (2018a) Identification of genes regulating ovary differentiation after pollination in hazel by comparative transcriptome analysis. BMC Plant Biol 18:84. https://doi.org/10.1186/s12870-018-1296-3
Cheng Y, Zhang L, Zhao Y et al (2018b) Analysis of SSR markers information and primer selection from transcriptome sequence of hybrid hazelnut Corylus heterophylla × C. avellana. Acta Hortic Sin 45:139–148. https://doi.org/10.16420/j.issn.0513-353x.2017-0281
Choi D, Kim JH, Lee Y (2008) Expansins in plant development. Adv Bot Res 47:47–97. https://doi.org/10.1016/S0065-2296(08)00002-5
Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17:268–281. https://doi.org/10.1105/tpc.104.026971
George L, Shawn MR, Jeffrey FH et al (2008) The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol 146:716–728. https://doi.org/10.1104/pp.107.108118
Gismondi A, Marco GD, Delorenzo M et al (2015) Upgrade of Castanea sativa (Mill.) genetic resources by sequencing of barcode markers. J Genet 94:519–524. https://doi.org/10.1007/s12041-015-0557-2
Guo X, Xu J, Cui X, Chen H, Qi H (2017) iTRAQ-based protein profiling and fruit quality changes at different development stages of oriental melon. BMC Plant Biol 17(28):28. https://doi.org/10.1186/s12870-017-0977-7
Hérouart D, Van MM, Inzé D (1994) Developmental and environmental regulation of the Nicotiana plumbaginifolia cytosolic Cu/Zn-superoxide dismutase promoter in transgenic tobacco. Plant Physiol 104:873–880. https://doi.org/10.1104/pp.104.3.873
Hollingsworth PM, Forrest LL, Spouge JL et al (2009) A DNA barcode for land plants. Proc Natl Acad Sci U S A 106:12794–12797. https://doi.org/10.1073/pnas.0905845106
Igarashi Y, Yoshiba Y, Sanada Y et al (1997) Characterization of the gene for delta1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant Mol Biol 33:857–865. https://doi.org/10.1023/A:100570240
Isaacson T, Damasceno CM, Saravanan RS et al (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc 1:769–774. https://doi.org/10.1038/nprot.2006.102
Jacob Y, Bergamin E, Donoghue MT et al (2014) Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343:1249–1253. https://doi.org/10.1126/science.1248357
Jan A, Yang G, Nakamura H, Ichikawa H, Kitano H, Matsuoka M, Matsumoto H, Komatsu S (2004) Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol 136:3670–3681. https://doi.org/10.1104/pp.104.052274
Kim S, Mollet JC, Dong J, Zhang K, Park SY, Lord EM (2003) Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc Natl Acad Sci U S A 100:16125–16130. https://doi.org/10.1073/pnas.2533800100
Kruger NJ (1994) The Bradford method for protein quantitation. In: Walker JM (ed) Methods in molecular biology. Humana Press, Totowa, pp 9–15. https://doi.org/10.1385/0-89603-268-X:9
Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG, Haag JD, Gould MN, Stewart RM, Kendziorski C (2013) EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29:1035–1043. https://doi.org/10.1093/bioinformatics/btt087
Li J, Yu M, Geng L et al (2010) The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J 64:482–497. https://doi.org/10.1111/j.1365-313X.2010.04344.x
Liu J, Cheng Y, Yan K, Liu Q, Wang Z (2012) The relationship between reproductive growth and blank fruit formation in Corylus heterophylla Fisch. Sci Hortic 136:128–134. https://doi.org/10.1016/j.scienta.2012.01.008
Liu J, Cheng Y, Liu C, Zhang C, Wang Z (2013) Temporal changes of disodium fluorescein transport in hazelnut during fruit development stage. Sci Hortic 150:348–353. https://doi.org/10.1016/j.scienta.2012.12.001
Liu J, Zhang H, Cheng Y, Kafkas S, Güney M (2014a) Pistillate flower development and pollen-tube growth mode during the delayed fertilization stage in Corylus heterophylla Fisch. Plant Reprod 27:145–152. https://doi.org/10.1007/s00497-014-0248-9
Liu J, Zhang H, Cheng Y, Wang J, Zhao Y, Geng W (2014b) Comparison of ultrastructure, pollen tube growth pattern and starch content in developing and abortive ovaries during the progamic phase in hazel. Front Plant Sci 5:528. https://doi.org/10.3389/fpls.2014.00528
Liu J, Xing J, Fang J, Ai P, Cheng Y (2018) New insight into ovary abortion during ovary development of hazelnut through a combined proteomic and transcriptomic analysis. Sci Hortic 234:36–48. https://doi.org/10.1016/j.scienta.2018.02.003
Lupini A, Araniti F, Sunseri F, Abenavoli MR (2014) Coumarin interacts with auxin polar transport to modify root system architecture in Arabidopsis thaliana. Plant Growth Regul 74:23–31. https://doi.org/10.1007/s10725-014-9893-0
Maniga A, Ghisaura S, Perrotta L, Marche MG, Cella R, Albani D (2017) Distinctive features and differential regulation of the DRTS genes of Arabidopsis thaliana. PLoS One 12:e0179338. https://doi.org/10.1371/journal.pone.0179338
McDowell JM, An YQ, Huang S et al (1996) The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol 111:699–711. https://doi.org/10.1104/pp.111.3.699
Molassiotis A, Tanou G, Filippou P, Fotopoulos V (2013) Proteomics in the fruit tree science arena: new insights into fruit defense, development, and ripening. Proteomics 13:1871–1884. https://doi.org/10.1002/pmic.201200428
Nilo R, Saffie C, Lilley K, Baeza-Yates R, Cambiazo V, Campos-Vargas R, González M, Meisel LA, Retamales J, Silva H, Orellana A (2010) Proteomic analysis of peach fruit mesocarp softening and chilling injury using difference gel electrophoresis (DIGE). BMC Genomics 11:43. https://doi.org/10.1186/1471-2164-11-43
Ogiwara H, Ui A, Kawashima S, Kugou K, Onoda F, Iwahashi H, Harata M, Ohta K, Enomoto T, Seki M (2007) Actin-related protein ARP4 functions in kinetochore assembly. Nucleic Acids Res 35:3109–3117. https://doi.org/10.1093/nar/gkm161
Ouyang J, Shao X, Li J (2000) Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J 24:327–334. https://doi.org/10.1046/j.1365-313x.2000.00883.x
Pascual MB, El-Azaz J, Torre FNDL et al (2016) Biosynthesis and metabolic fate of phenylalanine in conifers. Front Plant Sci 7:1030. https://doi.org/10.3389/fpls.2016.01030
Qin G, Meng X, Wang Q, Tian S (2009) Oxidative damage of mitochondrial proteins contributes to fruit senescence: a redox proteomics analysis. J Proteome Res 8:2449–2462. https://doi.org/10.1021/pr801046m
Reddy AS, Day IS (2001) Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol 2:1–19. https://doi.org/10.1186/gb-2001-2-7-research0024
Robinson SP, Jacobs AK, Dry IB (1997) A class IV chitinase is highly expressed in grape berries during ripening. Plant Physiol 114:771–778. https://doi.org/10.1104/pp.114.3.771
Schiøtt M, Romanowsky SM, Bækgaard L et al (2004) A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc Natl Acad Sci U S A 101:9502–9507. https://doi.org/10.1073/pnas.0401542101
Seberg O, Petersen G (2009) How many loci does it take to DNA barcode a crocus? PLoS One 4:e4598. https://doi.org/10.1371/journal.pone.0004598
Shahidi F, Alasalvar C, Liyana-Pathirana CM (2007) Antioxidant phytochemicals in hazelnut kernel (Corylus avellana L.) and hazelnut byproducts. J Agric Food Chem 55:1212–1220. https://doi.org/10.1021/jf062472o
Shen WH, Parmentier Y, Hellmann H, Lechner E, Dong A, Masson J, Granier F, Lepiniec L̈, Estelle M, Genschik P (2002) Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell 13:1916–1928. https://doi.org/10.1091/mbc.e02-02-0077
Song J, Du L, Li L et al (2015) Targeted quantitative proteomic investigation employing multiple reaction monitoring on quantitative changes in proteins that regulate volatile biosynthesis of strawberry fruit at different ripening stages. J Proteome 126:288–295. https://doi.org/10.1016/j.jprot.2015.06.004
Sudre D, Gutierrez-Carbonell E, Lattanzio G, Rellán-Álvarez R, Gaymard F, Wohlgemuth G, Fiehn O, Álvarez-Fernández A, Zamarreño AM, Bacaicoa E, Duy D, García-Mina JM, Abadía J, Philippar K, López-Millán AF, Briat JF (2013) Iron-dependent modifications of the flower transcriptome, proteome, metabolome, and hormonal content in an Arabidopsis ferritin mutant. J Exp Bot 64:2665–2688. https://doi.org/10.1093/jxb/ert112
Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, Lehesranta S, Mähönen AP, Kim JY, Jokitalo E, Sauer N, Scheres B, Nakajima K, Carlsbecker A, Gallagher KL, Helariutta Y (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21:1144–1155. https://doi.org/10.1016/j.devcel.2011.10.006
Wei Y, Xu H, Diao L, Zhu Y, Xie H, Cai Q, Wu F, Wang Z, Zhang J, Xie H (2015) Protein repair L-isoaspartyl methyltransferase 1 (PIMT1) in rice improves seed longevity by preserving embryo vigor and viability. Plant Mol Biol 89:475–492. https://doi.org/10.1007/s11103-015-0383-1
Wen B, Zhou R, Feng Q, Wang Q, Wang J, Liu S (2014) Iquant: an automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 14:2280–2285. https://doi.org/10.1002/pmic.201300361
Wolf C, Hennig M, Romanovicz D, Steinebrunner I (2007) Developmental defects and seedling lethality in apyrase AtAPY1 and AtAPY2 double knockout mutants. Plant Mol Biol 64:657–672. https://doi.org/10.1007/s11103-007-9184-5
Xu XM, Zhao Q, Rodrigo-Peiris T, Brkljacic J, He CS, Muller S, Meier I (2008) RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc Natl Acad Sci U S A 105:18637–18642. https://doi.org/10.1073/pnas.0806157105
Yun Z, Shuai J, Ding Y et al (2012) Comparative transcriptomics and proteomics analysis of citrus fruit, to improve understanding of the effect of low temperature on maintaining fruit quality during lengthy post-harvest storage. J Exp Bot 63:2873–2893. https://doi.org/10.1093/jxb/err390
Zang L, Zheng T, Chu Y, Ding C, Zhang W, Huang Q, Su X (2015) Genome-wide analysis of the fasciclin-like arabinogalactan protein gene family reveals differential expression patterns, localization, and salt stress response in populus. Front Plant Sci 6:1140. https://doi.org/10.3389/fpls.2015.01140
Zhu W, Huang CL, Zhang XH et al (2008) Analysis of ECA1 function by RNA interference in Arabidopsis thaliana. Plant Physiol Commun 44:469–475. https://doi.org/10.13592/j.cnki.ppj.2008.03.009
Zik M, Irish VF (2003) Flower development: initiation, differentiation, and diversification. Annu Rev Cell Dev Biol 19:119–140. https://doi.org/10.1146/annurev.cellbio.19.111301.134635
Data archiving statement
The data used for analysis in this study was provided as supplementary files of Figs. S1–S3 and Tables S1–S11.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31670681; 31770723) and Jilin Provincial Department of Education (No. JJKH20191012KJ; JJKH20190996KJ).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. M. Dandekar
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Figure 1
Protein identification overview (PNG 58 kb)
Supplementary Figure 2
Quantitation of chalcone synthase (CHS) and apyrase 1 (APY1) by western blot at three developmental stages. A: Quantitation of CHS in Fig. 9; B: Quantitation of APY1 in Fig. 9. The relative ratios of the signal intensity between target proteins (CHS and APY1) and internal reference β-actin were quantified. β-actin was used for normalization. Error bar represents the SE of three independent measurements. Different lowercase letters above each column denote significant differences determined by Student’s t test at the 5% level of significance. (JPG 29 kb)
Supplementary Figure 3
Original pictures of western blot at three developmental stages. Note: Set 1 to set 3 are three replicates of the western blot. (JPG 60 kb)
Supplementary Table 1
Detailed information of protein identification (XLSX 204 kb)
Supplementary Table 2
DEPs in S-vs-F paired comparison (XLSX 70 kb)
Supplementary Table 3
DEPs in T-vs-S paired comparison (XLSX 18 kb)
Supplementary Table 4
DEPs in T-vs-F paired comparison (XLS 3578 kb)
Supplementary Table 5
FPKM for 15 randomly chosen DEPs (XLSX 304 kb)
Supplementary Table 6
236 DEGs which were simultaneously differentially expressed at mRNA level (XLSX 142 kb)
Supplementary Table 7
FPKM for all identified unigenes (XLSX 565 kb)
Supplementary Table 8
FPKM for all identified DEGs (XLSX 12 kb)
Supplementary Table 9
FPKM for all identified proteins (XLSX 27 kb)
Supplementary Table 10
FPKM for all identified DEPs (XLSX 6264 kb)
Supplementary Table 11
FPKM for all interest proteins (XLSX 1073 kb)
Rights and permissions
About this article
Cite this article
Cheng, Y., Mou, Y., Zhang, X. et al. iTRAQ protein profiling reveals candidate proteins regulating ovary and ovule differentiation in pistillate inflorescences after pollination in hazel. Tree Genetics & Genomes 15, 21 (2019). https://doi.org/10.1007/s11295-019-1328-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-019-1328-7