Abstract
Pyrus pyrifolia is considered one of the most important cultivated Pyrus species. Hundreds of landraces and bred cultivars have been developed through the natural and artificial hybridizations necessary due to self-incompatibility. In this study, the genetic diversity of 478 Pyrus accessions, including Chinese landraces, bred cultivars, and wild samples, as well as introduced pear cultivars from Japan and Korea, was investigated with a set of 17 simple sequence repeat (SSR) markers distributed across all 17 linkage groups of the pear genome. A total of 121 alleles were detected, including 4 rare alleles with a frequency lower than 5%. Diversity statistics indicated a high level of genetic variation as quantified by the average values of the observed heterozygosity, the expected heterozygosity, and Wright’s fixation index, at 0.76, 0.78, and 0.02, respectively. Population structure and discriminant analysis of principal component analysis implied extensive genetic communication between sand pears in China and revealed four contiguous geographical clusters with overlapping geographical regions. The diversity of the four clusters and approximate Bayesian computation (ABC) indicated that sand pear spread from west to east along the Pearl River and Yangtze River valleys. High diversity and polyphyletic genetic components of cultivars in southwestern China further support southwestern China as the probable center of divergence for Pyrus species. A core collection of 80 out of 470 cultivars was selected, accounting for about 17% of accessions, and capturing 91% of all alleles, including all rare alleles. Our research provides a comprehensive understanding of sand pear germplasm in East Asia and constructs a preliminary core collection, which will be useful for association genetics studies, germplasm conservation, and breeding programs.




Similar content being viewed by others
References
Aranzana MJ, Abbassi E, Howad W, Arús P (2010) Genetic variation, population structure and linkage disequilibrium in peach commercial varieties. BMC Genet 11(1):69
Balas FC, Osuna MD, Domínguez G, Pérez-Gragera F, López-Corrales M (2014) Ex situ conservation of underutilised fruit tree species: establishment of a core collection for Ficus carica L. using microsatellite markers (SSRs). Tree Genet Genomes 10:703–710
Baraket G, Chatti K, Saddoud O, Abdelkarim AB, Mars M, Trifi M, Hannachi AS (2011) Comparative assessment of SSR and AFLP markers for evaluation of genetic diversity and conservation of fig, Ficus carica L., genetic resources in Tunisia. Plant Mol Biol Report 29:171–184
Beaumont MA, Zhang W, Balding DJ (2002) Approximate Bayesian computation in population genetics. Genetics 162:2025
Brown A (1989) Core collections: a practical approach to genetic resources management. Genome 31:818–824
Brown. A., Marshall, DR. (1973) Optimum sampling strategies in genetic conservation (summary). Lecture Notes in Computer Science 4768:36–56
Campbell CS, Evans RC, Morgan DR, Dickinson TA, Arsenault MP (2007) Phylogeny of subtribe Pyrinae (formerly the Maloideae, Rosaceae): limited resolution of a complex evolutionary history. Plant Syst Evol 266:119–145
Campoy JA, Ruiz D, Egea J, Rees DJG, Celton JM, Martínez-Gómez P (2011) Inheritance of flowering time in apricot (Prunus armeniaca L.) and analysis of linked quantitative trait loci (QTLs) using simple sequence repeat (SSR) markers. Plant Mol Biol Report 29:404–410
Challice JS, Westwood MN (1973) Numerical taxonomic studies of the genus Pyrus using both chemical and botanical characters. Bot J Linn Soc 67:121–148
Chen H, Song Y, Li L, Khan MA, Li X, Korban SS, Wu J, Zhang S (2015) Construction of a high-density simple sequence repeat consensus genetic map for pear (Pyrus spp.). Plant Mol Biol Report 33:316–325
Cornuet (2010) Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinformatics 11:401
Cornuet JM, Santos F, Beaumont MA, Robert CP, Marin JM, Balding DJ, Guillemaud T, Estoup A (2008) Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24:2713–2719
Dong D, Fu X, Yuan F, Chen P, Zhu S, Li B, Yang Q, Yu X, Zhu D (2014) Genetic diversity and population structure of vegetable soybean (Glycine max (L.) Merr.) in China as revealed by SSR markers. Genet Resour Crop Evol 61:173–183
Dos Santos ARF, Ramos-Cabrer AM, Díaz-Hernández MB, Pereira-Lorenzo S (2011) Genetic variability and diversification process in local pear cultivars from northwestern Spain using microsatellites. Tree Genet Genomes 7:1041–1056
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater JM, Zyprian E, Moreira FM (2013) Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol 13(1):39
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fagundes NJR, Ray N, Beaumont M, Neuenschwander S, Salzano FM, Bonatto SL, Excoffier L (2007) Statistical evaluation of alternative models of human evolution. Proceedings of the National Academy of Sciences 104(45):17614–17619.
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fan L, Zhang M, Liu Q, Li L, Song Y, Wang L, Zhang S, Wu J (2013) Transferability of newly developed pear SSR markers to other Rosaceae species. Plant Mol Biol Report 31:1271–1282
Franco J, Crossa J, Taba S, Shands H (2005) A sampling strategy for conserving genetic diversity when forming core subsets. Crop Sci 45:1035–1044
Hamrick JL, Godt M (1996) Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society B-Biological Sciences 351:1291–1298
Hoelzel AR (2010) Looking backwards to look forwards: conservation genetics in a changing world. Conserv Genet 11:655–660
Hokanson SC, Lamboy WF, Szewc-McFadden AK, McFerson JR (2001) Microsatellite (SSR) variation in a collection of Malus (apple) species and hybrids. Euphytica 118:281–294
Hu J, Wang P, Su Y, Wang R, Li Q, Sun K (2015) Microsatellite diversity, population structure, and core collection formation in melon germplasm. Plant Mol Biol Report 33:439–447
Jiang S, Zong Y, Yue X, Postman J, Teng Y, Cai D (2015) Prediction of retrotransposons and assessment of genetic variability based on developed retrotransposon-based insertion polymorphism (RBIP) markers in Pyrus L. Mol Gen Genomics 290:225–237
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Kim K, Chung H, Cho G, Ma K, Chandrabalan D, Gwag J, Kim T, Cho E, Park Y (2007) PowerCore: a program applying the advanced M strategy with a heuristic search for establishing core sets. Bioinformatics 23:2155–2162
Lassois L, Denancé C, Ravon E, Guyader A, Guisnel R, Hibrand-Saint-Oyant L, Poncet C, Lasserre-Zuber P, Feugey L, Durel C (2016) Genetic diversity, population structure, parentage analysis, and construction of core collections in the French apple germplasm based on SSR markers. Plant Mol Biol Report 34:827–844
Le Cunff L, Fournier-Level A, Laucou V, Vezzulli S, Lacombe T, Adam-Blondon A, Boursiquot J, This P (2008) Construction of nested genetic core collections to optimize the exploitation of natural diversity in Vitis vinifera L. subsp. sativa. BMC Plant Biol 8(1):31
Lin Z, Zhang X, Nie Y, He D, Wu M (2003) Construction of a genetic linkage map for cotton based on SRAP. Chin Sci Bull 48:2064–2068
Liu J, Zheng X, Potter D, Hu C, Teng Y (2012) Genetic diversity and population structure of Pyrus calleryana (Rosaceae) in Zhejiang province, China. Biochem Syst Ecol 45:69–78
Liu J, Sun P, Zheng X, Potter D, Li K, Hu C, Teng Y (2013) Genetic structure and phylogeography of Pyrus pashia L. (Rosaceae) in Yunnan Province, China, revealed by chloroplast DNA analyses. Tree Genet Genomes 9:433–441
Liu QW, Song Y, Liu L, Zhang MY, Sun JM, Zhang SL, Wu J (2015) Genetic diversity and population structure of pear (Pyrus spp.) collections revealed by a set of core genome-wide SSR markers. Tree Genetics & Genomes 11(6):1–22
Lo EY, Stefanović S, Christensen KI, Dickinson TA (2009) Evidence for genetic association between East Asian and western North American Crataegus L. (Rosaceae) and rapid divergence of the eastern North American lineages based on multiple DNA sequences. Mol Phylogenet Evol 51:157–168
Lombard, P.B., Westwood, M.N., 1987. Pear rootstocks
Lu J, Wu J, Zhang S, Wu H, Zhang Y (2011) Genetic diversity and polygentic relationship among pears reveled by SSR analysis. Journal of Nanjing Agricultural University 2:009
Newton AC, Allnutt TR, Gillies A, Lowe AJ, Ennos RA (1999) Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14:140–145
Odong TL, Jansen J, Van Eeuwijk FA, van Hintum TJ (2013) Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation. Theor Appl Genet 126:289–305
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pu F, Wang Y (1963) Pomology of China. Pears. Shanghai Scientific and Technical Publishers, Shanghai (in Chinese)
Richards CM, Volk GM, Reilley AA, Henk AD, Lockwood DR, Reeves PA, Forsline PL (2009) Genetic diversity and population structure in Malus sieversii, a wild progenitor species of domesticated apple. Tree Genet Genomes 5:339–347
Rubtsov GA (1944) Geographical distribution of the genus Pyrus and trends and factors in its evolution. Am Nat 78:358–366
Schoen DJ, Brown AH (1993) Conservation of allelic richness in wild crop relatives is aided by assessment of genetic markers. Proc Natl Acad Sci 90:10623–10627
Shen D, Bo W, Xu F, Wu R (2014) Genetic diversity and population structure of the Tibetan poplar (Populus szechuanica var. tibetica) along an altitude gradient. BMC Genet 15:1
Slatkin M (1985) Rare alleles as indicators of gene flow. Evolution 39:53–65
Song Y, Fan L, Chen H, Zhang M, Ma Q, Zhang S, Wu J (2014) Identifying genetic diversity and a preliminary core collection of Pyrus pyrifolia cultivars by a genome-wide set of SSR markers. Sci Hortic 167:5–16
Taberlet P, Fumagalli L, Wustsaucy AG, Cosson JF (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Tautz D (1989) Hypervariabflity of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463–6471
Teng Y, Tanabe K, Tamura F, Itai A (2002) Assessment of genetic relatedness among large-fruited pear cultivars native to east asia using rapd markers. Acta Horticulturae (587):139–145
Vargas AM, de Andrés MT, Ibáñez J (2016) Maximization of minority classes in core collections designed for association studies. Tree Genet Genomes 12
Vercken E, Fontaine MC, Gladieux P, Hood ME, Jonot O, Giraud T (2010) Glacial refugia in pathogens: European genetic structure of anther smut pathogens on Silene latifolia and Silene dioica. PLoS Pathog 6:e1001229
Wu J, Li L, Li M, Khan MA, Li X, Chen H, Yin H, Zhang S (2014) High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. Journal of Experimental Botany 65(20):5771–5781
Wuyun T, Amo H, Xu J, Ma T, Uematsu C, Katayama H (2015) Population structure of and conservation strategies for wild Pyrus ussuriensis Maxim. in China. PLoS One 10:e0133686
Yeh FC, Yang R, Boyle TB, Ye ZH, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular biology and biotechnology centre. University of Alberta, Canada, p 10
Yu (1979a) Taxonomy of the fruit tree in China (in Chinese). Beijing, China Agr Press
Yu DJ (1979b) Pyrus pyrifolia. Flora Republicae Popularis Sinicae 36:365
Zhang P, Li J, Li X, Liu X, Zhao X, Lu Y (2011) Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers. PLoS One 6:e27565
Zhang MY, Fan L, Liu QZ, Song Y, Wei SW (2014) A novel set of EST-derived SSR markers for pear and cross-species transferability in Rosaceae. Plant Mol Biol Report 32:290-302
Zheng X, Cai D, Potter D, Postman J, Liu J, Teng Y (2014) Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol Phylogenet Evol 80:54–65
Zong Y, Sun P, Liu J, Yue X, Niu Q, Teng Y (2014) Chloroplast DNA-based genetic diversity and phylogeography of Pyrus betulaefolia (Rosaceae) in northern China. Tree Genet Genomes 10:739–749
Funding
This work was financially supported by the National Science Foundation of China (31672111), the Earmarked Fund for China Agriculture Research System (CARS-29), the Science Foundation for Distinguished Young Scientists in Jiangsu Province (BK20150025), and The Six Talent Peaks Project in Jiangsu Province (2014-NY-025).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Fig. S1
Distribution of pairwise relatedness coefficients among the P. pyrifolia individuals. rxy values among cultivars are normally distributed around a mean of zero, with a low variance between pairs of individuals. (PDF 121 kb)
Fig. S2
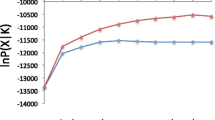
Modeling of number of genepools in P. pyrifolia using STRUCTURE. Ln P(D) and delta K, calculated according to Evanno et al. (2005), plotted against the number of modeled genepools (K). (JPG 170 kb)
Fig. S3
Inference of the number of clusters. Bayesian information criterion (BIC) is provided for different numbers of clusters (from 1 to 50). The chosen number of clusters (8) is crossed in black. (PDF 310 kb)
Fig. S4
Scatterplots of DAPC. Groups are shown by different colors and inertia ellipses; dots represent individual strains. (PDF 368 kb)
ESM 1
(DOCX 38.6 kb)
ESM 2
(DOCX 13.7 kb)
ESM 3
(DOCX 17.6 kb)
Rights and permissions
About this article
Cite this article
Xue, L., Liu, Q., Hu, H. et al. The southwestern origin and eastward dispersal of pear (Pyrus pyrifolia) in East Asia revealed by comprehensive genetic structure analysis with SSR markers. Tree Genetics & Genomes 14, 48 (2018). https://doi.org/10.1007/s11295-018-1255-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-018-1255-z