Abstract
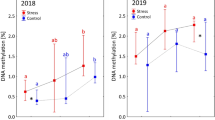
The role of epigenetic phenomena in plant adaptation is becoming widely recognized and the potential of epigenetics for forestry practice has been demonstrated as well. In this study, methylation-sensitive amplification polymorphism (MSAP) markers were investigated in 20 European beech (Fagus sylvatica L.) provenances that cover most of Europe and were planted in two climatically contrasting provenance trial plots. Correlations of cytosine-methylation patterns at five loci and overall DNA methylation with climatic conditions of the sites of population origin and budburst phenology were detected, suggesting that methylation at particular loci was influenced by the weather or photoperiod during embryogenesis or even earlier. Alternation of methylation patterns may also have been caused by genetic mutation. Frequencies of methylation patterns at three loci differed between the two trial locations, indicating that a climatically induced change of methylation during the ontogeny occurs as well. The results suggest that the rules for collection, transfer, and use of forest reproductive materials should also consider epigenetic effects.
Similar content being viewed by others
References
Alonso C, Pérez R, Bazaga P, Medrano M, Herrera CM (2015) MSAP markers and global cytosine methylation in plants: a literature survey and comparative analysis for wild-growing species. Mol Ecol Resour 16:80–90. https://doi.org/10.1111/1755-0998.12426
Angers B, Castonguay E, Massicotte R (2010) Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol Ecol 19:1283–1295. https://doi.org/10.1111/j.1365-294X.2010.04580.x
Ariani A, Romeo S, Groover AT, Sebastiani L (2016) Comparative epigenomic and transcriptomic analysis of Populus roots under excess Zn. Environ Exp Bot 132:16–27. https://doi.org/10.1016/j.envexpbot.2016.08.005
Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427:164–167. https://doi.org/10.1038/nature02269
Bossdorf O, Richards CL, Pigliucci M (2008) Epigenetics for ecologists. Ecol Lett 11:106–115. https://doi.org/10.1111/j.1461-0248.2007.01130.x
Bräutigam K, Vining KJ, Lafon-Placette C, Fossdal CG, Mirouze M, Marcos JG, Fluch S, Fraga MF, Guevara MÁ, Abarca D, Johnsen Ø, Maury S, Strauss SH, Campbell MM, Rohde A, Díaz-Sala C, Cervera MT (2013) Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol Evol 3:399–415. https://doi.org/10.1002/ece3.461
Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM (2011) DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 6:e14524. https://doi.org/10.1371/journal.pone.0014524
Cubas P, Vincent C, Coen E (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401:157–161
Doyle JJ, Doyle JL (1987) Isolation of DNA from fresh plant tissue. Phytochem Bull 19:11–15
Dubin MJ, Zhang P, Meng D, Remigereau M-S, Osborne EJ, Casale FP, Drewe P, Kahles A, Jean G, Vilhjálmsson B, Jagoda J, Irez S, Voronin V, Song Q, Long Q, Rätsch G, Stegle O, Clark RM, Nordborg M (2015) DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 4:e05255. https://doi.org/10.7554/eLife.05255
Feng S, Cokus SJ, Zhang X, Chen PJ, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107:8689–8694. https://doi.org/10.1073/pnas.1002720107
Fraga MF, Rodriguez R, Canal MJ (2002) Genomic DNA methylation-demethylation during aging and reinvigoration of Pinus radiata. Tree Physiol 22:813–816. https://doi.org/10.1093/treephys/22.11.813
Fulneček J, Kovařík A (2014) How to interpret methylation sensitive amplified polymorphism (MSAP) profiles? BMC Genet 15:2. https://doi.org/10.1186/1471-2156-15-2
Fulneček J, Matyášek R, Kovařík A (2002) Distribution of 5-methylcytosine residues in 5S rRNA genes in Arabidopsis thaliana and Secale cereale. Mol Gen Genomics 268:510–517. https://doi.org/10.1007/s00438-002-0761-7
Gömöry D, Paule L (2011) Trade-off between height growth and spring flushing in common beech (Fagus sylvatica L.) Ann For Sci 68:975–984. https://doi.org/10.1007/s13595-011-0103-1
Gömöry D, Ditmarová L, Hrivnák M, Jamnická G, Kmet’ J, Krajmerová D, Kurjak D (2015) Differentiation in phenological and physiological traits in European beech (Fagus sylvatica L.). Eur J For Res 134:1075–1085
Gouil Q, Baulcombe DC (2016) DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet 12:e1006526. https://doi.org/10.1371/journal.pgen.1006526
Gugger PF, Fitz-Gibbon S, Pellegrini M, Sork VL (2016) Species-wide patterns of DNA methylation variation in Quercus lobata and their association with climate gradients. Mol Ecol 25:1665–1680. https://doi.org/10.1111/mec.13563
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978. https://doi.org/10.1002/joc.1276
Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324:1451–1454. https://doi.org/10.1126/science.1172417
Johnsen Ø (1989) Phenotypic changes in progenies of northern clones of Picea abies (L.) Karst. grown in a southern seed orchard. I. Frost hardiness in a phytotron experiment. Scand J Forest Res 4:317–330. https://doi.org/10.1080/02827588909382569
Johnsen Ø, Dæhlen OG, Østreng G, Skrøppa T (2005) Daylength and temperature during seed production interactively affect adaptive performance of Picea abies progenies. New Phytol 168:589–596. https://doi.org/10.1111/j.1469-8137.2005.01538.x
Joost S, Bonin A, Bruford MW, Després L, Conord C, Erhardt G, Taberlet P (2007) A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol Ecol 16:3955–3969. https://doi.org/10.1111/j.1365-294X.2007.03442.x
Kawakatsu T, Huang SSC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He YP, Chen HM, Dubin M, Lee CR, Wang CM, Bemm F, Becker C, O'Neil R, O'Malley RC, Quarless DX, Schork NJ, Weigel D, Nordborg M, Ecker JR, 1001 Genomes Consortium (2016) Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166:492–505. https://doi.org/10.1016/j.cell.2016.06.044
Kawashima T, Berger F (2014) Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet 15:613–624. https://doi.org/10.1038/nrg3685
Kovařík A, Koukalová B, Bezděk M, Opatrný Z (1997) Hypermethylation of tobacco heterochromatic loci in response to osmotic stress. Theor Appl Genet 95:301–306. https://doi.org/10.1007/s001220050563
Krajmerová D, Hrivnák M, Ditmarová Ľ, Jamnická G, Kmeť J, Kurjak D, Gömöry D (2017) Nucleotide polymorphisms associated with climate, phenology and physiological traits in European beech (Fagus sylvatica L.) New For 48:463–477 https://doi.org/10.1007/s11056-017-9573-9
Labra M, Ghiani A, Citterio S, Sgorbati S, Sala F, Vannini C, Ruffini-Castiglione M, Bracale M (2002) Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol 4:694–699. https://doi.org/10.1055/s-2002-37398
Magri D, Vendramin GG, Comps B, Dupanloup I, Geburek T, Gömöry D, Latalowa M, Litt T, Paule L, Roure JM, Tantau I, van der Knaap WO, Petit RJ, de Beaulieu J-L (2006) A new scenario for the Quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytol 171:199–221
Maunakea AK, Chepelev I, Cui K, Zhao K (2013) Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res 23:1256–1269. https://doi.org/10.1038/cr.2013.110
Paun O, Bateman RM, Fay MF, Hedrén M, Civeyrel L, Chase MW (2010) Stable epigenetic effects impact adaptation in allopolyploid orchids (Dactylorhiza: Orchidaceae). Mol Biol Evol 27:2465–2473. https://doi.org/10.1093/molbev/msq150
Pigliucci M (1996) How organisms respond to environmental changes: from phenotypes to molecules (and vice versa). Trends Ecol Evol 11:168–173. https://doi.org/10.1016/0169-5347(96)10008-2
Raj S, Bräutigam K, Hamanishi ET, Wilkins O, Schroeder W, Mansfield SD, Plant AL, Campbell MM (2011) Clone history shapes Populus drought responses. Proc Natl Acad Sci USA 108:12521–12526. https://doi.org/10.1073/pnas.1103341108
Reyna-López GE, Simpson J, Ruiz-Herrera J (1997) Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol Gen Genet 253:703–710. https://doi.org/10.1007/s004380050374
Richards CL, Schrey AW, Pigliucci M (2012) Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecol Lett 15:1016–1025. https://doi.org/10.1111/j.1461-0248.2012.01824.x
Richards CL, Alonso C, Becker C et al. (2017) Ecological plant epigenetics: evidence from model and non-model species, and the way forward. bioRxiv. https://www.biorxiv.org/content/ early/2017/04/25/130708. Accessed 15 July 2017. doi: https://doi.org/10.1101/130708
Rohde A, Junttila O (2008) Remembrances of an embryo: long-term effects on phenology traits in spruce. New Phytol 177:2–5. https://doi.org/10.1111/j.1469-8137.2007.02319.x
Santamaria ME, Hasbun R, Valera MJ (2009) Acetylated H4 histone and genomic DNA methylation patterns during bud set and bud burst in Castanea sativa. J Plant Physiol 166:1360–1369. https://doi.org/10.1016/j.jplph.2009.02.014
Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ, Ecker JR (2013) Patterns of population epigenomic diversity. Nature 495:193–198. https://doi.org/10.1038/nature11968
Schrey AW, Alvarez M, Foust CM, Kilvitis HJ, Lee JD, Liebl AL, Martin LB, Richards CL, Robertson M (2013) Ecological epigenetics: beyond MS-AFLP. Integr Comp Biol 53:340–350. https://doi.org/10.1093/icb/ict012
Schulz B, Eckstein RL, Durka W (2013) Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol Ecol Resour 13:642–653. https://doi.org/10.1111/1755-0998.12100
Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowitz W (2008) MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15:259–267. https://doi.org/10.1038/nsmb.1391
Skrøppa T, Tollefsrud MM, Sperisen C, Johnsen Ø (2009) Rapid change in adaptive performance from one generation to the next in Picea abies—Central European trees in a Nordic environment. Tree Genet Genomes 6:93–99. https://doi.org/10.1007/s11295-009-0231-z
Su C, Wang C, He L, Yang C, Wang Y (2014) Shotgun bisulfite sequencing of the Betula platyphylla genome reveals the tree's DNA methylation patterning. Int J Mol Sci 15:22874–22886. https://doi.org/10.3390/ijms151222874
Vergeer P, Wagemaker N, Ouborg NJ (2012) Evidence for an epigenetic role in inbreeding depression. Biol Lett 8:798–801. https://doi.org/10.1098/rsbl.2012.0494
von Wühlisch G, Krusche D, Muhs HJ (1995) Variation in temperature sum requirement for flushing of beech provenances. Silvae Genet 44:343–346
von Wühlisch G, Liesebach M, Muhs HJ, Stephan BR (1998) A network of international beech provenance trials. In: Turok J, Kremer A, de Vries SM (eds) First EUFORGEN meeting on social broadleaves. International Plant Genetic Resources Institute, Rome, pp 164–172
Walder RY, Langtimm CJ, Chatterjee R, Walder JA (1983) Cloning of the MspI modification enzyme. The site of modification and its effects on cleavage by MspI and HpaII. J Biol Chem 258:1235–1241
Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y (2010) DNA methylation mediated by a microRNA pathway. Mol Cell 38:465–475. https://doi.org/10.1016/j.molcel.2010.03.008
Yakovlev IA, Fossdal CG, Johnsen Ø (2010) MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytol 187:1154–1169. https://doi.org/10.1111/j.1469-8137.2010.03341.x
Yakovlev IA, Lee Y, Rotter B, Olsen JE, Skrøppa T, Johnsen Ø, Fossdal CG (2014) Temperature-dependent differential transcriptomes during formation of an epigenetic memory in Norway spruce embryogenesis. Tree Genet Genomes 10:355–366. https://doi.org/10.1007/s11295-013-0691-z
Yakovlev IA, Carneros E, Lee Y, Olsen JE, Fossdal CG (2016) Transcriptional profiling of epigenetic regulators in somatic embryos during temperature induced formation of an epigenetic memory in Norway spruce. Planta 243:1237–1249. https://doi.org/10.1007/s00425-016-2484-8
Zeng FS, Zhou S, Zhan YG, Dong J (2014) Drought resistance and DNA methylation of interspecific hybrids between Fraxinus mandshurica and Fraxinus americana. Trees 28:1679–1692. https://doi.org/10.1007/s00468-014-1077-z
Acknowledgements
The provenance experiment has been established through the realization of the project European Network for the Evaluation of the Genetic Resources of Beech for Appropriate Use in Sustainable Forestry Management (AIR3-CT94-2091) under the coordination of H.-J. Muhs and G. von Wühlisch. The experimental plots Tále and Zbraslav were established by L. Paule and V. Hynek, respectively. The study was supported by a research grant of the Slovak Research and Development Agency APVV-0135-12 and the project of the Ministry of Agriculture of the Czech Republic—Resolution RO0117 (reference number 6779/2017-MZE-14151.).We also thank to K. Willingham for linguistic correction.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Data Archiving Statement
MSAP profile data have been deposited at the TreeGenes Data Repository (dendrome.ucdavis.edu/treegenes/) under the accession number TGDR069.
Additional information
Communicated by G. G. Vendramin
Rights and permissions
About this article
Cite this article
Hrivnák, M., Krajmerová, D., Frýdl, J. et al. Variation of cytosine methylation patterns in European beech (Fagus sylvatica L.). Tree Genetics & Genomes 13, 117 (2017). https://doi.org/10.1007/s11295-017-1203-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1203-3