Abstract
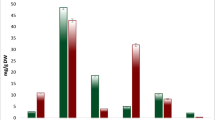
Paulownia australis has important economic and ecological values. In this study, the morphological and physiological changes of the leaves in diploid and autotetraploid P. australis under salt stress were analyzed. To detect related genes and gain a comprehensive perspective on the molecular mechanisms underlying salt tolerance in P. australis, transcriptome-wide gene expression profiling was conducted in the leaves of the diploid and autotetraploid P. australis under control and salinity conditions, respectively. Evaluation of the responses against salinity stress revealed the superiority of autotetraploid over diploid in terms of salinity tolerance. Changes in physiological parameters in diploid P. australis (PA2) and tetraploid P. australis (PA4) plants in response to salt stress were measured. Transcriptome data revealed that many of the common unigenes which were involved in accumulation of compatible solutes, oxidative stress detoxification, ion homeostasis, and signal transduction showed significant differences between the two accessions in response to salt stress. A number of salt-responsive unigenes were identified in two accessions of P. australis under salt stress. Furthermore, the differentially expressed unigenes found to be common in both accessions may be useful genetic resources for further genetic improvement of Paulownia using transgenic approaches.






Similar content being viewed by others
References
Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N (2000) Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol 123:1047–1056
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Armitage P, Berry G, Matthews JNS (2008) Statistical methods in medical research. Blackwell, London
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1
Asano T, Hakata M, Nakamura H, Aoki N, Komatsu S, Ichikawa H, Hirochika H, Ohsugi R (2011) Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol Biol 75:179–191
Ashraf M, Harris P (2004) Potential biochemical indicators of salinity tolerance in plants. Plant Science 166:3–16
Barrs H, Weatherley P (1962) A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci 15:413–428
Batelli G, Verslues P, Agius F, Qiu Q, Fujii H, Pan S, Schumaker K, Grillo S, Zhu J (2007) SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol Cell Biol 27:7781–7790
Bates L, Waldren R, Teare I (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188
Bieniawska Z, Paul Barratt DH, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49:810–828
Bohnert HJ, Nelson DE, Jensen RG (1995) Adaptations to environmental stresses. Plant Cell 7:1099
Bojović B, Stojanović J (2005) Chlorophyll and carotenoid content in wheat cultivars as a function of mineral nutrition. Arch Biol Sci 57:283–290
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chao DY, Dilkes B, Luo H, Douglas A, Yakubova E, Lahner B, Salt DE (2013) Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341:658–659
Cheng N-H, Pittman JK, Zhu JK, Hirschi KD (2004) The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem 279:2922–2926
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Davies JP, Yildiz FH, Grossman AR (1999) Sac3, an Snf1-like serine/threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11:1179–1190
Dhindsa RS, Plumb-Dhindsa P, Thorpe TA (1981) Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J Exp Bot 32:93–101
Dong Y, Fan G, Zhao Z, Deng M (2014) Compatible solute, transporter protein, transcription factor, and hormone-related gene expression provides an indicator of drought stress in Paulownia fortunei. Funct Integr Genomics 14:479–491
Fan G, Wei Z, Yang Z (2009) Induction of autotetraploid of Paulownia australis and its in vitro plantlet regeneration. Journal of Northwest A & F University 37:83–90.
Forrest KL, Bhave M (2007) Major intrinsic proteins (MIPs) in plants: a complex gene family with major impacts on plant phenotype. Funct Integr Genomics 7:263–289
Gong Z, Koiwa H, Cushman MA, Ray A, Bufford D, Kore-eda S, Matsumoto TK, Zhu J, Cushman JC, Bressan RA (2001) Genes that are uniquely stress regulated in salt overly sensitive (SOS) mutants. Plant Physiol 126:363–375
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Guy C, Haskell D, Neven L, Klein P, Smelser C (1992) Hydration-state-responsive proteins link cold and drought stress in spinach. Planta 188:265–270
Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Biol 51:463–499
Hashimoto M, Komatsu K, Maejima K, Okano Y, Shiraishi T, Ishikawa K, Takinami Y, Yamaji Y, Namba S (2012) Identification of three MAPKKKs forming a linear signaling pathway leading to programmed cell death in Nicotiana benthamiana. BMC Plant Biol 12:1
Hodges DM, DeLong JM, Forney CF, Prange RK (1999) Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207:604–611
Hong Z, Lakkineni K, Zhang Z, Verma DPS (2000) Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol 122:1129–1136
Hsu S, Hsu Y, Kao C (2003) The effect of polyethylene glycol on proline accumulation in rice leaves. Biol Plant 46:73–78
Huang Z, Zhao L, Chen D, Liang M, Liu Z, Shao H, Long X (2013) Salt stress encourages proline accumulation by regulating proline biosynthesis and degradation in Jerusalem artichoke plantlets. PLoS One 8:e62085
Irigoyen J, Einerich D, Sánchez-Díaz M (1992) Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativd) plants. Physiol Plant 84:55–60
Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 99:138–148
Joung J-G, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, Fei Z (2009) Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol 151:1758–1768
Kanehisa M, Araki M, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Kerepesi I, Galiba G (2000) Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Science 40:482–487
Kishor PK, Sangam S, Amrutha RN, Laxmi PS, Naidu KR, Rao K, Rao S, Reddy KJ, Theriappan P, Sreenivasulu N (2005) Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Curr Sci 88:424–438
Kolukisaoglu Ü, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134:43–58
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma 12:1
Li W, Zhang C, Lu Q, Wen X, Lu C (2011) The combined effect of salt stress and heat shock on proteome profiling in Suaeda salsa. J Plant Physiol 168:1743–1752
Liu F, Guo Y, Gu D, Xiao G, Chen Z, Chen S (1996) Salt tolerance of transgenic plants with BADH cDNA. Acta Genet Sin 24:54–58
Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci 97:3730–3734
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150:244–256
Mahajan S, Pandey GK, Tuteja N (2008) Calcium-and salt-stress signaling in plants: shedding light on SOS pathway. Arch Biochem Biophys 471:146–158
Meiri D, Breiman A (2009) Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90. 1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J 59:387–399
Miller G, Suzuki N, CIFTCI-YILMAZ S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33:453–467
Molina C, Zaman-Allah M, Khan F, Fatnassi N, Horres R, Rotter B, Steinhauer D, Amenc L, Drevon J, Winter P (2011) The salt-responsive transcriptome of chickpea roots and nodules via deepSuperSAGE. BMC Plant Biol 11:1
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999) Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett 461:205–210
Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147:1251–1263
Ottow EA, Brinker M, Teichmann T, Fritz E, Kaiser W, Brosché M, Kangasjärvi J, Jiang X, Polle A (2005) Populus euphratica displays apoplastic sodium accumulation, osmotic adjustment by decreases in calcium and soluble carbohydrates, and develops leaf succulence under salt stress. Plant Physiol 139:1762–1772
Panikulangara TJ, Eggers-Schumacher G, Wunderlich M, Stransky H, Schöffl F (2004) Galactinol synthase1. A novel heat shock factor target gene responsible for heat-induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiol 136:3148–3158
Peng Z, He S, Gong W, Sun J, Pan Z, Xu F, Lu Y, Du X (2014) Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genomics 15:1
Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci 99:8436–8441
Qiu JL, Qiu J, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, MacKinlay J, Loake GJ, Mundy J, Morris PC (2008) Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol 148:212–222
Rahman S, Miyake H, Takeoka Y (2002) Effects of exogenous glycinebetaine on growth and ultrastructure of salt-stressed rice seedlings (Oryza sativa L.) Plant Prod Sci 5:33–44
Rahman H, Jagadeeshselvam N, Valarmathi R, Sachin B, Sasikala R, Senthil N, Sudhakar D, Robin S, Muthurajan R (2014) Transcriptome analysis of salinity responsiveness in contrasting genotypes of finger millet (Eleusine coracana L.) through RNA-sequencing. Plant Mol Biol 85:485–503
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Romeis T, Ludwig AA, Martin R, Jones JD (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20:5556–5567
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Seki M, Umezawa T, Urano K, Shinozaki K (2007) Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10:296–302
Sun Y, Oberley LW, Li Y (1988) A simple method for clinical assay of superoxide dismutase. Clin Chem 34:497–500
Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought-and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29:417–426
Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu J, Shinozaki K (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135:1697–1709
Tan W, Blake TJ, Boyle TJ (1992) Drought tolerance in faster-and slower-growing black spruce (Picea mariana) progenies: I. Stomatal and gas exchange responses to osmotic stress. Physiol Plant 85:639–644
Tan FQ, Tu H, Liang WJ, Long JM, Wu XM, Zhang HY, Guo WW (2015) Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. junos cv. Ziyang xiangcheng) suggests its potential value for stress resistance improvement. BMC Plant Biol 15:89
Tang RJ, Liu H, Bao Y, Lv QD, Yang L, Zhang HX (2010) The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol Biol 74:367–380
Tang S, Liang H, Yan D, Zhao Y, Han X, Carlson JE, Xia X, Yin W (2013) Populus euphratica: the transcriptomic response to drought stress. Plant Mol Biol 83:539–557
Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15:141–152
Tuteja N, Mahajan S (2007) Calcium signaling network in plants: an overview. Plant Signal Behav 2:79–85
Tyerman S, Niemietz C, Bramley H (2002) Plant aquaporins: multifunctional water and solute channels with expanding roles. Plant Cell Environ 25:173–194
Urano K, Kurihara Y, Seki M, Shinozaki K (2010) ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr Opin Plant Biol 13:132–138
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132
Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X (2005) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139:822–835
Wang Y, Ying J, Kuzma M, Chalifoux M, Sample A, McArthur C, Uchacz Y, Sarvas C, Wan J, Dennis DT (2005) Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J 43:413–424
Wang J, Zhu XP, Gao R, Lin CL, Li Y, Xu QC, Piao CG, Li XD, Li HF, Tian GZ (2010) Genetic and serological analyses of elongation factor EF-Tu of Paulownia witches’-broom phytoplasma (16SrI-D). Plant Pathol 59:972–981
Welch WJ, Brown CR (1996) Influence of molecular and chemical chaperones on protein folding. Cell Stress Chaperones 1:109
Wu T, Kong XP, Zong XJ, Li DP, Li DQ (2011) Expression analysis of five maize MAP kinase genes in response to various abiotic stresses and signal molecules. Mol Biol Rep 38:3967–3975
Wu J, Wang J, Pan C, Guan X, Wang Y, Liu S, He Y, Chen J, Chen L, Lu G (2014) Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS One 9:e103032
Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14:S165–S183
Xu E, Fan G, Niu S, Zhao Z, Deng M, Dong Y (2015) Transcriptome sequencing and comparative analysis of diploid and autotetraploid Paulownia australis. Tree Genet Genomes 11:1–13
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006) The regulatory domain of SRK2E/OST1/SnRK2. 6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281:5310–5318
Zhang X, Zhai X, Fan G, Deng M, Zhao Z (2013) Observation on microstructure of leaves and stress tolerance analysis of different tetraploid Paulownia. J Henan Agric Univ 46:646–650
Zhang J, Feng J, Lu J, Yang Y, Zhang X, Wan D, Liu J (2014) Transcriptome differences between two sister desert poplar species under salt stress. BMC Genomics 15:1
Funding information
This work was supported by the Joint Funds of the National Natural Science Foundation of China (NSFC) (Grant No. U1204309), the Fund of the Transformation Project of the National Agricultural Scientific and Technological Achievement of China (Grant No. 2012GB2D000271), the Central Financial Forestry Science Promotion Project (Grant No. GTH [2012]01), the Fund of the Science Key Program of Department of Henan Education (Grant No. 12A220003), and the Fund of the Technology Innovation Team Project of Zhengzhou (Grant No. 121PCXTD515).
Author information
Authors and Affiliations
Contributions
Guoqiang Fan conceived and designed the experiments, and supervised the study. Yanpeng Dong performed experiments, analyzed data, and wrote the manuscript. Yanpeng Dong revised the manuscript. Zhenli Zhao and Enkai Xu performed the experiments. Minjie Deng, Limin Wang and Suyan Niu contributed reagents or other essential material.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Data archiving statement
Sequence data from this article have been deposited with the National Center for Biotechnology Information Sequence Read Archive database (http://www.ncbi. nlm.nih.gov/sra) under accession no. SRP059262.
Additional information
Communicated by W.-W. Guo
Electronic supplementary material
ESM 1
The process of the de novo transcriptome assembly. (GIF 34 kb)
ESM 2
Primers of quantitative RT-PCR analysis of candidate salt response genes. -f represents forward primers and -r represents reverse primers. (DOCX 17 kb)
ESM 3
Distribution of unigene lengths in the transcriptome of P. australis. The sizes of all unigenes were calculated. (GIF 256 kb)
ESM 4
Unigene annotations based on their BLASTX hits in the Nr, KEGG, Swiss-Prot, KOG, and GO database. (XLSX 2645 kb)
ESM 5
Classification of the Karyotic Orthologous Groups (KOG) for the transcriptome of P. australis (GIF 16 kb)
ESM 6
Classification of the gene ontology (GO) for the transcriptome of P. australis (GIF 1439 kb)
ESM 7
KEGG annotation of unigenes (XLSX 121 kb)
ESM 8
Co-regulated differentially expressed unigenes in both the PA2T vs. PA2W and PA4T vs. PA4W comparisons. PA2T, 15 days salt-treated diploid; PA2W, well-watered diploid; PA4T, 15 days salt-treated tetraploid; PA4W, well-watered tetraploid. (XLSX 739 kb)
ESM 9
Co-regulated differentially expressed unigenes in both the PA4W vs. PA2W and PA4T vs. PA2T comparisons. PA4W, well-watered tetraploid; PA2W, well-watered diploid; PA4T, 15 days salt-treated tetraploid; PA2T, 15 days salt-treated diploid. (XLSX 699 kb)
ESM 10
Co-regulated differentially expressed unigenes in the four libraries. PA2T, 15 days salt-treated diploid; PA2W, well-watered diploid; PA4T, 15 days salt-treated tetraploid; PA4W, well-watered tetraploid. (XLSX 167 kb)
ESM 11
DEUs function in the accumulation of compatible solutes, oxidative stress detoxification, ion homeostasis and salt stress in the discussion section. (XLSX 130 kb)
ESM 12
KEGG pathway analysis results for the co-regulated differentially expressed unigenes in the four libraries.PA2T, 15 days salt-treated diploid; PA2W, well-watered diploid; PA4T, 15 days salt-treated tetraploid; PA4W, well-watered tetraploid. (XLSX 15 kb)
Rights and permissions
About this article
Cite this article
Dong, Y., Fan, G., Zhao, Z. et al. Transcriptome-wide profiling and expression analysis of two accessions of Paulownia australis under salt stress. Tree Genetics & Genomes 13, 97 (2017). https://doi.org/10.1007/s11295-017-1179-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1179-z