Abstract
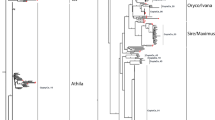
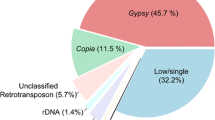
The availability of nearly complete moso bamboo genome sequences permits the detailed discovery and cross-species comparison of transposable elements (TEs) between Bambusoideae and other Poaceae species at the whole genome level. Long terminal repeat retroelements (LTR-retroelements) are the single largest components of most plant genomes and can substantially impact the genome in various ways. Through a combination of structure- and homology-based approaches, we initially investigated 982 LTR-retroelement families comprising 2,004,644 LTR-retroelement sequences, which accounted for more than 40% of the moso bamboo genome. Further analysis revealed that the ratio of solo LTRs to intact elements (S/I) in moso bamboo is significantly low (approximately 0.28:1), indicating that bamboo LTR-retroelements might have undergone relatively low frequencies of unequal recombination and illegitimate recombination. Phylogenetic analysis revealed four Ty1-copia and five Ty3-gypsy evolutionary lineages that were present before the divergence of eudicot and monocot species, but the scales and timeframes within which they proliferated significantly varied across families and lineages. Insertion time estimates showed that LTR-retroelements were amplified for approximately 0~3 million years and had longer periods of activity than those of rice and Arabidopsis. These findings suggest that the expansion of LTR-retroelements might be responsible for host large genome size during moso bamboo evolution.



Similar content being viewed by others
References
Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, Bennetzen JL (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5:e1000732
Bennetzen JF (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Devos KM, Brown JK, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Du J, Tian Z, Hans CS, Laten HM, Cannon SB, Jackson SA, Shoemaker RC, Ma J (2010) Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J 63:584–598
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Eickbush TH, Jamburuthugoda VK (2008) The diversity of retroelements and the properties of their reverse transcriptases. Virus Res 134:221–234
Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Fu J (2001) Chinese moso bamboo: its importance. Bamboo 22:5–7
Gorinsek B, Gubensek F, Kordis D (2004) Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol 21:781–798
Gui YJ, Zhou Y, Wang Y et al (2010) Insights into the bamboo genome: syntenic relationships to rice and sorghum. J Integr Plant Biol 52:1008–1015
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol 5:225
Havecker ER, Gao X, Voytas DF (2005) The Sireviruses, a plant-specific lineage of the Ty1/copia retrotransposons, interact with a family of proteins related to dynein light chain. Plant Physiol 139:857–868
Hoen DR, Hickey G, Bourque G, Casacuberta J, Cordaux R, Feschotte C, Fiston-Lavier AS, Hua-Van A, Hubley R, Kapusta A, Lerat E, Maumus F, Pollock DD, Quesneville H, Smit A, Wheeler TJ, Bureau TE, Blanchette M (2015) A call for benchmarking transposable element annotation methods. Mob DNA 6:1–9
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110(1-4):462–467
Kashkush K, Khasdan V (2007) Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes. Genetics 177:1975–1985
Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33:102–106
Kimura M, Ota T (1972) On the stochastic model for estimation of mutational distance between homologous proteins. J Mol Evol 2:87–90
Llorens C, Muñoz-Pomer A, Bernad L, Botella H, Moya A (2009) Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol Direct 4:41
Llorens C, Futami R, Covelli L et al (2011) The gypsy database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res 39:70–74
Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A 101:12404–12410
Ma J, Jackson SA (2006) Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res 16:251–259
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
McCarthy EM, McDonald JF (2003) LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19:362–367
Paterson AH, Bowers JE, Bruggmann R et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Peng Z, Lu Y, Li L et al (2013) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat Genet 45:456–461
Piegu B, Guyot R, Picault N et al (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269
Platt RN, Blanco-Berdugo L, Ray DA (2016) Accurate transposable element annotation is vital when analyzing new genome assemblies. Genome Biology and Evolution 8(2):403–410
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Steinbauerova V, Neumann P, Novak P, Macas J (2011) A widespread occurrence of extra open reading frames in plant Ty3/gypsy retrotransposons. Genetica 139(11–12):1543–1555
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Bio Evol 30:2725–2729
Tian Z, Rizzon C, Du J, Zhu L, Bennetzen JL, Jackson SA, Gaut BS, Ma J (2009) Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res 19:2221–2230
Wang H, Liu JS (2008) LTR retrotransposon landscape in Medicago truncatula: more rapid removal than in rice. BMC Genomics 9:382
Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17:1072–1081
Wicker T, Sabot F, Hua-Van A et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Wright DA, Voytas DF (2002) Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res 12:122–131
Zhao H, Peng Z, Fei B, Li L, Hu T, Gao Z, Jiang Z (2014) BambooGDB: a bamboo genome database with functional annotation and an analysis platform. Database Vol article ID:bau006
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Authors’ contribution
M. B. Zhou—designation of the experiments, identification and classification of LTR-retroelement, and writing of paper; B. J. Hu, Y. H. Zhu—estimation of LTR-retroelement insertion date; all authors read and approved the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
Funding the Talents Program of the Natural Science Foundation of Zhejiang Province (Grant No. LR12C16001), and The National Natural Science Foundation of China (Grant Nos. 31470615 and 31270645) supported this study.
Data Archiving Statement
Genomic sequences and gene annotation information of moso bamboo presented in this report are available in the bamboo genome database (BambooGDB, http://www.bamboogdb.org/index.jsp). The plant representative elements of four lineages of Ty1-Copia (Sire, Oryco, Retrofit, and Tork) and five lineages of Ty3-Gypsy (Crm, Del, Reina, Athila, and Tat) are available in the Gypsy Database (GyDB) (http://www.gydb.org/index.php/Main_Page). All full-length sequences of Ty1-Copia and Ty3-gypsy retroelements of A. thaliana and O. sativa are available in the Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/index.jsp) and Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/home_contacts.shtml), respectively.
Additional information
Communicated by C. Chen
Electronic supplementary material
Supplementary file S1
An overview of the workflow for the identification of moso bamboo LTR-retroelements. (DOC 37 kb)
Supplementary table S2
Representative sequence of each moso bamboo LTR-retroelement family, including element sizes, LTR sizes, TSD sizes, element sequences, LTR sequences, TSD sequences, primer binding site (PBS) sequences, and polypurine tract (PPT) sequences. (XLS 377 kb)
Rights and permissions
About this article
Cite this article
Zhou, M., Hu, B. & Zhu, Y. Genome-wide characterization and evolution analysis of long terminal repeat retroelements in moso bamboo (Phyllostachys edulis). Tree Genetics & Genomes 13, 43 (2017). https://doi.org/10.1007/s11295-017-1114-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1114-3