Abstract
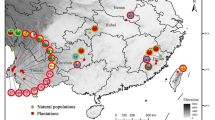
Persian walnut (Juglans regia L.) is the world’s most widely grown nut crop, but large-scale assessments and comparisons of the genetic diversity of the crop are notably lacking. To guide the conservation and utilization of Persian walnut genetic resources, genotypes (n = 189) from 25 different regions in 14 countries on three continents were sampled to investigate their genetic relationships and diversity using ten microsatellite (SSR) loci. The SSRs amplified from 3 to 25 alleles per locus, with a mean value of 11.5 alleles per locus. The mean values of observed and expected heterozygosity were 0.62 and 0.73, respectively. Based on Nei’s genetic identity, accessions from Bratislava (Slovakia) and Antalya (Turkey) showed the lowest similarity (0.36), while accessions from Algeria and Tunisia as well as accessions from Debrecen (Hungary) and Trnava (Slovakia) had the highest similarity (0.97). Two populations from Iran (Alborz and Ardabil) had the highest number of private alleles (7 and 5), but they were quite different as they also had the lowest genetic identity when compared to the remaining populations as well as to each other. Although overall differentiation among regions was relatively low (F st = 0.07), cluster analysis grouped accessions generally but not completely according to geography. STRUCTURE software confirmed these results and divided the accessions into two main groups, separating accessions collected from Europe and North Africa from those from Greece and the Near East. Results indicate the presence of a likely center of diversity for Persian walnut in Eastern and Southeastern Europe. They also provide information that can be used to devise conservation actions. Notably, the genetic diversity of threatened populations from two regions in Iran should be conserved.





Similar content being viewed by others
References
Addisalem AB, Bongers F, Kassahun T, Smulders MJM (2016) Genetic diversity and differentiation of the frankincense tree (Boswellia papyrifera (Del.) Hochst) across Ethiopia and implications for its conservation. For Ecol Manag 360:253–260. doi:10.1016/j.foreco.2015.10.038
Aradhya M, Woeste K, Velasco D (2010) Genetic diversity, structure and differentiation in cultivated walnut (Juglans regia L). Acta Hort 861:127–132
Avanzato D, McGranahan GH, Vahdati K, Botu M, Iannamico L, Van Assche J (2014) Following walnut footprints (Juglans regia L.) cultivation and culture, folklore and history, traditions and uses. Scripta Horticulturae, p 442
Bayazit S, Kazan K, Gülbitti S, Çevik V, Ayanoğlu H, Ergül A (2006) AFLP analysis of genetic diversity in low chill requiring walnut (Juglans regia L.) genotypes from Hatay, Turkey. Sci Hort 111:394–398
Christopoulos MV, Rouskas D, Tsantili E, Bebeli PJ (2010) Germplasm diversity and genetic relationships among walnut (Juglans regia L.) cultivars and Greek local selections revealed by Inter-Simple Sequence Repeat (ISSR) markers. Sci Hort 125:584–592
Cornille A, Gladieux P, Smulders MJM, Roldán-Ruiz I, Laurens F, Le Cam B, Nersesyan A, Clavel J, Olonova M, Feugey L, Gabrielyan I, Zhang XG, Tenaillon MI, Giraud T (2012) New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet 8:e1002703. doi:10.1371/journal.pgen.1002703
Dangl GS, Woeste K, Aradhya MK, Koehmstedt A, Simon C, Potter D, Leslie CA, McGranaham G (2005) Characterization of 14 microsatellite markers for genetic analysis and cultivar identification of walnut. J Amer Sco Hort Sci 130:348–354
Dogan Y, Kafkas S, Sutyemez M, Turemis N (2014) Assessment and characterization of genetic relationships of walnut (Juglans regia L.) genotypes by three types of molecular markers. Sci Hort 168:81–87
Domoto PA (2002) Walnut tree named ‘Domoto’. United States Plant Patent № US PP12, 898 P2
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. doi:10.1007/s12686-011-9548-7
Ebrahimi A, Vahdati K, Fallahi E (2007) Improved success of Persian walnut grafting under environmentally controlled conditions. Int J Fruit Sci 6(4):3–12
Ebrahimi A, Fatahi R, Zamani Z (2011) Analysis of genetic diversity among some Persian walnut genotypes (Juglans regia L.) using morphological traits and SSRs markers. Sci Hort 130:146–151. doi:10.1016/j.scienta.2011.06.028
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Foroni I, Rao R, Woeste K, Gallitelli M (2005) Characterization of Juglans regia L. through SSR markers and evaluation of genetic relationships among cultivars and the ‘Sorrento’ landrace. J Hortic Sci Biotechnol 80:49–53
Foroni I, Woeste K, Monti LM, Rao R (2007) Identification of ‘Sorrento’ walnut using simple sequence repeats (SSRs). Genet Resour Crop Evol 54:1081–1094. doi:10.1007/s10722-006-9187-0
Germain, E (2004) Inventory of walnut research, germplasm and references. FAO Regional Office For Europe, REU Technical Series 66, Rome, Italy, p 264
Jafari Sayadi MH, Vahdati K, Mozafari J, Mohajer MRM, Leslie CA (2012) Natural hyrcanian population of Persian walnut (Juglans regia L.) in Iran. Acta Hortic 948:97–101. doi:10.17660/ActaHortic.2012.948.10
Kalinowski ST (2005) HP-rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5:187–189
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
McGranahan GH, Charles A, Leslie CA, Philips HA, Dandaker A (1998) Walnut propagation. In: Ramos D (ed) Walnut production manual. University of California, DANR Publ, Davis, pp. 71–83
Mitra SK, Rathore DS, Bose TK (1991) Walnut. Temperate fruits, vol 27. Published by Horticulture and Allied Publishers, Calcutta, pp. 377–414
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nicese FP, Hormasa JI, McGranaham GH (1998) Molecular characterization and genetic relatedness among walnut (Juglans regia L.) genotypes based on RAPD markers. Euphytica 101:199–206
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pollegioni P, Woeste K, Olimpieri I, Marandola D, Cannata F, Malvolti ME (2011) Long term human impacts on genetic structure of Italian walnut inferred by SSR markers. Tree Genet Genomes 7:707–723
Pollegioni P, Woeste KE, Chiocchini F, Olimpieria L, Tortolano V, Clark J, Hemery GE, Mapelli S, Malvoti ME (2014) Landscape genetics of Persian walnut (Juglans regia L.) across its Asian range. Tree Genet Genomes 10:1027–1043
Pollegioni P, Woeste K, Chiocchini F, Del Lugo S, Cannata F, Olimpieri I, Tortolano V, Clark J, Hemery GE, Mapelli S, Malvolti EM (2015) Ancient humans influenced the current spatial genetic structure of common [Persian] walnut populations in Asia. PLoS One 7:707–723. doi:10.137/journal.pone.0135980
Pop IL, Vicol AC, Botu M, Raica PA, Vahdati K, Pamfil D (2013) Relationships of walnut cultivars in a germplasm collection: comparative analysis of phenotypic and molecular data. Sci Hort 153:124–135
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2012) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Topcu H, Coban N, Woeste K, Sutyemez M, Kafkas S (2015) Developing new microsatellite markers in walnut (Juglans regia L.) from Juglans nigra genomic GA enriched library. EKIN 1-2:93–99
Vahdati K, Mohseni Pourtaklu S, Karimi R, Barzehkar R, Amiri R, Mozaffari M, Woeste K (2015) Genetic diversity and gene flow of some Persian walnut populations in southeast of Iran revealed by SSR markers. Plant Syst Evol 301:691–699. doi:10.1007/s00606-014-1107-8
Wang H, Pei D, Gu R, Wang B (2008) Genetic diversity and structure of walnut populations in central and southwestern China revealed by microsatellite markers. J Am Soc Hortic Sci 133:197–203
Watanabe S, Noma N, Nishida T (2016) Flowering phenology and mating success of the heterodichogamous tree Machilus thunbergii Sieb. et Zucc (Lauraceae). Plant Species Biology 31:29–37
Woeste K, Burns R, Rhodes O, Michler C (2002) Thirty polymorphic nuclear microsatellite loci from black walnut. J Hered 93:58–60
Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Edmonton, Alberta, Canada
Zhao P, Zhang S, Woeste K (2012) Genotypic data changes family rank for growth and quality traits in a black walnut (Juglans nigra L.) progeny test. New For. doi:10.1007/s11056-012-9343-7
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Data archiving
If the manuscript is accepted, genotype data will be submitted to TreeGenes database.
Disclaimer
Mention of a trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the U.S. Dept. of Agriculture and does not imply its approval to the exclusion of other products or vendors that also may be suitable.
Additional information
Communicated by A.M. Dandekar
Aziz Ebrahimi and Abdolkarim Zarei contributed equally.
Electronic supplementary material
ESM 1
(DOCX 247 kb)
Rights and permissions
About this article
Cite this article
Ebrahimi, A., Zarei, A., Lawson, S. et al. Genetic diversity and genetic structure of Persian walnut (Juglans regia) accessions from 14 European, African, and Asian countries using SSR markers. Tree Genetics & Genomes 12, 114 (2016). https://doi.org/10.1007/s11295-016-1075-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1075-y