Abstract
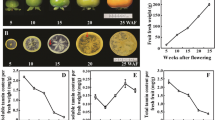
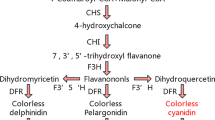
Russet skin is a very important trait that allows pear fruits to defend themselves against biotic and abiotic stresses. Small RNAs from a russet skin mutant ‘Xiusu’ derived from a ‘Dangshansuli’ pear were sequenced by high-throughput sequencing to reveal the role of miRNAs in the regulation of pear russet skin formation. A total of 12,158,547 and 12,053,678 high-quality reads were obtained for ‘Dangshansuli’ and ‘Xiusu’, respectively, with the majority between 19 and 25 nt in size. Forty-four and 45 known miRNAs were identified in the ‘Dangshansuli’ and ‘Xiusu’ libraries, respectively, and these miRNAs belonged to 31 miRNA families. The expression levels of 534 miRNAs varied drastically, ranging from 0 to 493,274 reads with a logarithm of fold changes between −9.33 and 12.71. In addition, 215 and 228 novel miRNAs with high-abundance were detected in ‘Dangshansuli’ and ‘Xiusu’, respectively. Many miRNAs, especially miR396, miR408, and the novel miRNAs, miR102, miR274, miR42, and miR442, were potentially involved in suberin biosynthesis and showed differential expression between the exocarp of ‘Dangshansuli’ and that of ‘Xiusu’. The relative expression levels of known and novel miRNAs as determined by quantitative PCR indicated that those miRNAs may contribute to the formation of mutant russet pear fruit skin.


Similar content being viewed by others
References
Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16:2463–2480
Albert Z, Ivanics B, Molnaŕ A, Miskó A, Tóth M, Papp I (2013) Candidate genes of cuticle formation show characteristic expression in the fruit skin of apple. Plant Growth Regul 70:71–78
Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36:1282–1290
An FM, Hsiao SR, Chan MT (2011) Sequencing-based approaches reveal low ambient temperature-responsive and tissue-specific microRNAs in Phalaenopsis orchid. PLoS One 6:e18937
Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB (2007) The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19:351–368
Bernards MA (2002) Demystifying suberin. Can J Bot 80:227–240
Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301:336–338
Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10:673–683
Chong YT, Gidda SK, Sanford C, Parkinson J, Mullen RT, Goring DR (2010) Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol 185:401–419
Ding YF, Zhu C (2009) The role of microRNAs in copper and cadmium homeostasis. Biochem Bioph Res Co 386:6–10
Eldem V, Celikkol Akcay U, Ozhuner E, Bakır Y, Uranbey S, Unver T (2012) Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS One 7:e50298
Gidda SK, Shockey JM, Rothstein SJ, Dyer JM, Mullen RT (2009) Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol Bioch 47:867–879
Go YS, Kim H, Kim HJ, Suh MC (2014) Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26:1666–1680
Graça J (2010) Hydroxycinnamates in suberin formation. Phytochem Rev 9:85–91
Griffiths-Jones S (2006) miRBase: the microRNA sequence database. Methods Mol Biol 342:129–138
Hála M, Cole R, Synek L, Drdová E, Pečenková T, Nordheim A, Lamkemeyer T, Madlung J, Hochholdinger F, Žárský V (2008) An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell 20:1330–1345
Heng W, Liu L, Wang MD, Jia B, Liu P, Ye ZF, Zhu LW (2014) Differentially expressed genes related to the formation of russet fruit skin in a mutant of ‘Dangshansuli’ pear (Pyrus bretchnederi Rehd.) determined by suppression subtractive hybridization. Euphytica 196:285–297
Huppi K, Volfovsky N, Mackiewicz M, Runfola T, Jones TL, Martin SE, Stephens R, Caplen NJ (2007) MicroRNAs and genomic instability. Semin Cancer Biol 17:65–73
Inoue M, Chiang SH, Chang L, Chen XW, Saltiel AR (2006a) Compartmentalization of the exocyst complex in lipid rafts controls Glut4 vesicle tethering. Mol Biol Cell 17:2303–2311
Inoue E, Kasumi M, Sakuma F, Anzai H, Amano K, Hara H (2006b) Identification of RAPD marker linked to fruit skin color in Japanese pear (Pyrus pyrifolia Nakai). Sci Hortic-Amsterdam 107:254–258
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Khanal BP, Grimm E, Knoche M (2012) Russeting in apple and pear: a plastic periderm replaces a stiff cuticle. AoB PLANTS 5:pls048
Kuo HF, Chiou TJ (2011) The role of microRNAs in phosphorus deficiency signaling. Plant Physiol 156:1016–1024
Legay S, Guerriero G, Deleruelle A, Lateur M, Evers D, André CM, Hausman JF (2015) Apple russeting as seen through the RNA-seq lens: strong alterations in the exocarp cell wall. Plant Mol Biol 88:21–40
Li BS, Qin YR, Duan H, Yin WL, Xia XL (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot 62:3765–3779
Liu HH, Tian X, Li YJ, CA W, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Liu J, Dyer D, Wang J, Wang S, Du X, Xu B, Zhang H, Wang X, Hu W (2013a) 3-oxoacyl-ACP reductase from Schistosoma japonicum: integrated in silico-in vitro strategy for discovering antischistosomal lead compounds. PLoS One 8:e64984
Liu S, Yang H, Zhao J, Zhang YH, Song AX, Hu HY (2013b) NEDD8 ultimate buster-1 long (NUB1L) protein promotes transfer of NEDD8 to proteasome for degradation through the P97UFD1/NPL4 complex. J Biol Chem 288:31339–31349
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402–408
Lu XY, Huang XL (2008) Plant miRNAs and abiotic stress responses. Biochem Bioph Res Co 368:458–462
Luo X, Gao Z, Shi T, Cheng Z, Zhang Z, Ni Z (2013) Identification of miRNAs and their target genes in peach (Prunus persica L.) using high-throughput sequencing and degradome analysis. PLoS One 8:e79090
Morozova O, Marra MA (2008) Applications of next-generation sequencing technologies in functional genomics 92:255–264
Niu Q, Qian M, Liu G, Yang F, Teng Y (2013) A genome-wide identification and characterization of mircoRNAs and their targets in ‘Suli’ pear (Pyrus pyrifolia white pear group. Planta 238:1095–1112
O'Hara P, Slabas AR, Fawcett T (2007) Antisense expression of 3-oxoacyl-ACP reductase affects whole plant productivity and causes collateral changes in activity of fatty acid synthase components. J Biol Chem 48:736–744
Pollard M, Beisson F, Li Y, Ohlrogge JB (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13:236–246
Qin YR, Duan ZX, Xia XL, Yin WL (2011) Expression profiles of precursor and mature microRNAs under dehydration and high salinity shock in Populus euphratica. Plant Cell Rep 30:1893–1907
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16:1616–1626
Schmidt HW, Schönherr J (1982) Fine structure of isolated and non-isolated potato tuber periderm. Planta 154:76–80
Song C, Fang J, Li X, Liu H, Thomas Chao C (2009) Identification and characterization of 27 conserved microRNAs in citrus. Planta 230:671–685
Song C, Wang C, Zhang C, Kibet Korir N, Yu H, Ma Z, Fang J (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11:431
Sugar D, Basile SR (2008) Color and russet variation among selections of ‘Bosc’ pear. J Am Pomol Soc 62:77–81
Varkonyi-Gasic E, Gould N, Sandanayaka M, Sutherland P, MacDiarmid RM (2010) Characterisation of micrornas from apple (Malus domestica ‘Royal Gala’) vascular tissue and phloem sap. BMC Plant Biol 10:159
Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X (2011) miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot 62:761–773
Wang T, Pan H, Wang J, Yang W, Cheng T, Zhang Q (2014) Identification and profiling of novel and conserved microRNAs during the flower opening process in Prunus mume via deep sequencing. Mol Gen Genomics 289:169–183
Wu XM, Liu MY, Ge XX, Xu Q, Guo WW (2011) Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange. Planta 233:495–505
Wu J, Wang Z, Shi Z et al (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Wu J, Wang D, Liu Y, Wang L, Qiao X, Zhang S (2014) Identification of miRNAs involved in pear fruit development and quality. BMC Genomics 15:953
Xia R, Zhu H, An YQ, Beers EP, Liu ZR (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13:R47
Xu Q, Liu Y, Zhu A, Wu X, Ye J, Yu K, Guo W, Deng X (2010) Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics 11:246
Ye KY, Chen Y, Hu XW, Guo JC (2013) Computational identification of microRNAs and their targets in apple. Genes Genom 35:377–385
Yu X, Wang H, YZ L, de Ruiter M, Cariaso M, Prins M, van Tunen A, He YK (2012) Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot 63:1025–1038
Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang ZY (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42:689–707
Zhang BH, Pan XP, Cobb GP, Anderson TA (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289:3–16
Zhang J, Ai X, Guo W, Peng S, Deng X, Hu C (2012) Identification of miRNAs and their target genes using deep sequencing and degradome analysis in trifoliate orange [Poncirus trifoliate (L.) Raf]. Mol Biotechnol 51:44–57
Zhang X, Li X, Liu J (2014) Identification of conserved and novel cold-responsive microRNAs in trifoliate orange (Poncirus trifoliata (L.)Raf.) using high-throughput sequencing. Plant Mol Biol Rep 32:328–341
Zhao BT, Ge LF, Liang RQ, Li W, Ruan KC, Lin HX, Jin YX (2009) Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol 10:29
Zhou LG, Liu YH, Liu ZC, Kong DY, Duan M, Luo LJ (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61:4157–4168
Acknowledgments
This project was supported by the National Natural Science Foundation of China (31101519) and the earmarked fund for the China Agriculture Research System (CARS-29-14). We thank American Journal Experts for helpful suggestions and revision of the paper.
Author contribution statement
HW and ZLW conceived and designed the study. HHN and YJY collected and prepared RNA samples for transcriptome analysis. JB, WZT, and LP performed RNA extraction and qPCR. LL and YZF contributed to bioinformatic analysis. HW and JB contributed to writing the manuscript and data analysis. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Data archiving statement
The data sets supporting the results of this article (BioProject: PRJNA279180) are currently being submitted to the National Center for Biotechnology Information, and additional information will be added once available.
Additional information
Communicated by C. Dardick
Wei Heng and Bing Jia contributed equally to this work.
Rights and permissions
About this article
Cite this article
Heng, W., Jia, B., Huang, Hn. et al. Identification of russet-associated microRNAs in the exocarp of a Dangshansuli pear mutant (Pyrus bretschneideri Rehd.) by high-throughput sequencing. Tree Genetics & Genomes 12, 107 (2016). https://doi.org/10.1007/s11295-016-1058-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1058-z