Abstract
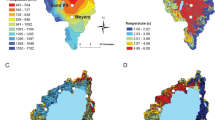
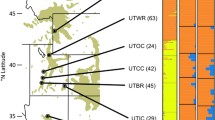
Demographic and environmental forces shape geographical patterns of genetic diversity. Knowledge thereof is not only important for evolutionary ecologists but, in light of future climate change, will be of interest to conservation biologists as well. Sugar pine (Pinus lambertiana Dougl.) is an ecologically important species found in mixed conifer forests across western North America. We applied a candidate-gene-based environmental study to infer spatial patterns in neutral genetic variation and to identify genetic variants associated with local adaptation to drought. Using a panel of 186 candidate gene single nucleotide polymorphisms (SNP), we genotyped 313 individual trees sampled across the entire state of California, USA. We found evidence for a large-scale subdivision into two genetic clusters along the latitudinal axis and increased genetic similarity among sugar pines within a 200–300-km boundary. Associating allelic to environmental variation indicated nine putative SNPs related to local adaptation to drought. These results provide insights into neutral population structure across the natural range of sugar pine and further substantiated a key role of the mitochondrial import inner membrane machinery in enhanced tolerance to drought and constitute important steps into unravelling the eco-evolutionary dynamics in sugar pine.




Similar content being viewed by others
References
Aitken SN, Whitlock MC (2013) Assisted gene flow to facilitate local adaptation to climate change. Annu Rev Ecol Evol Syst 44:367–388
Aitken SN, Yeaman S, Holliday JA, Wang TL, Curtis-McLane S (2008) Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol Appl 1:95–111
Angell N, Waring KM, Graves TA (2013) Predicting height growth of sugar pine regeneration using stand and individual tree characteristics. Forestry 1–13, doi:10.1093/forestry/cpt1028.
Berg JJ, Coop G (2014) A population genetic signal of polygenic adaptation. PLoS Genet 10:e1004412
Bigelow SW, North MP, Horwath WR (2009) Resource-dependent growth models for Sierran mixed-conifer saplings. Open Forest Sci J 2:31–40
Coop G, Witonsky D, Di Rienzo A, Pritchard JK (2010) Using environmental correlations to identify loci underlying local adaptation. Genetics 185:1411–1423
De Kort H, Vandepitte K, Bruun HH et al (2014) Landscape genomics and a common garden trial reveal adaptive differentiation to temperature across Europe in the tree species Alnus glutinosa. Mol Ecol 23:4709–4721
De Kort H, Vandepitte K, Mergeay J, Mijnsbrugge KV, Honnay O (2015) The population genomic signature of environmental selection in the widespread insect-pollinated tree species Frangula alnus at different geographical scales. Heredity 115:415–425
De La Torre AR, Wang T, Jaquish B, Aitken SN (2014) Adaptation and exogenous selection in a Picea glauca × Picea engelmannii hybrid zone: implications for forest management under climate change. New Phytol 201:687–699
Duncan O, Murcha MW, Whelan J (2013) Unique components of the plant mitochondrial protein import apparatus. Biochim Biophys Acta 2:304–313
Eckert A, Maloney P, Vogler D et al (2015) Local adaptation at fine spatial scales: an example from sugar pine (Pinus lambertiana, Pinaceae). Tree Genetics & Genomes 11:1–17
Eckert AJ, Bower AD, González-Martinez SC et al (2010) Back to nature: ecological genomics of loblolly pine (Pinus taeda, Pinaceae). Mol Ecol 19:3789–3805
Eckert AJ, Bower AD, Wegrzyn JL et al (2009) Asssociation genetics of coastal Douglas fir (Pseudotsuga menziesu var. menziesii, Pinaceae). I. Cold-hardiness related traits. Genetics 182:1289–1302
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Farjon A, Filer DL (2013) An atlas of the world’s conifers: an analysis of their distribution, biogeography, diversity and conservation status. Brill, Leiden
Ferrell GT (1996) The influence of insect pests and pathogens on Sierra forests. In: Sierra Nevada Ecosystem Project: Final Report to Congress (ed. Erman, D.C.). University of California, Centers for Water and Wildland Resources, Davis, pp 1177–1192
Fietto LG, Costa MDL, Cruz CD et al (2007) Identification and in silico analysis of the Citrus HSP70 molecular chaperone gene family. Genet Mol Biol 30:881–887
Forsythe WC, Rykiel EJ, Stahl RS, Wu HI, Schoolfield RM (1995) A model comparison for daylength as a function of latitude and day of year. Ecol Model 80:87–95
Frichot E, Schoville SD, Bouchard G, François O (2013) Testing for associations between loci and environmental gradients using latent factor mixed models. Mol Biol Evol 30:1687–1699
Friendly M (2002) Corrgrams: exploratory displays for correlation matrices. The American Statistician 56:316–324
Geraldes A, Farzaneh N, Grassa CJ et al (2014) Landscape genomics of Populus trichocarpa: the role of hybridization, limited gene flow, and natural selection in shaping patterns of population structure. Evolution 68:3260–3280
Guichoux E, Garnier-Gere P, Lagache L et al (2013) Outlier loci highlight the direction of introgression in oaks. Mol Ecol 22:450–462
Günther T, Coop G (2013) Robust Identification of Local Adaptation from Allele Frequencies. Genetics 195:205–220
Hijmans RJ, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:1–6
Jones MR, Forester BR, Teufel AI et al (2013) Integrating landscape genomics and spatially explicit approaches to detect loci under selection in clinal populations. Evolution 67:3455–3468
Kinloch BB (2003) White pine blister rust in North America: past and prognosis. Phytopathology 93:1044–1047
Kitzmiller JH (2004) Adaptive genetic variation in Sugar pine. In: Breeding and genetic resources of five-needle pines: growth, adaptability, and pest resistance (ed. Sniezko R). Proceedings Conference IUFRO Working Party 2.02.15. Ogden (UT): USDA Forest Service, Rocky Mountain Research Station. RMRS-P-32. p 103–123
Kremer A, Ronce O, Robledo-Arnuncio JJ et al (2012) Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecol Lett 15:378–392
Krzywinski M, Schein J, Birol I et al (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Kurz WA, Dymond CC, Stinson G et al (2008) Mountain pine beetle and forest carbon feedback to climate change. Nature 452:987–990
Laudenslayer WF, Darr HH (1990) Historical effects of logging on forests of the Cascade and Sierra Nevada ranges of California. Trans West Wildl Soc 26:12–23
Le Corre V, Kremer A (2012) The genetic differentiation at quantitative trait loci under local adaptation. Mol Ecol 21:1548–1566
Liston A, Parker-Defeniks M, Syring JV, Willyard A, Cronn R (2007) Interspecific phylogenetic analysis enhances intraspecific phylogeographical inference: a case study in Pinus lambertiana. Mol Ecol 16:3926–3937
Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4:981–994
Mokranjac D, Sichting M, Neupert W, Hell K (2003) Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J 22:4945–4956
Mosca E, Eckert AJ, Di Pierro EA et al (2012) The geographical and environmental determinants of genetic diversity for four alpine conifers of the European Alps. Mol Ecol 21:5530–5545
Mote P, Salathé E Jr (2010) Future climate in the Pacific Northwest. Clim Change 102:29–50
Namroud MC, Beaulieu J, Juge N, Laroche J, Bousquet J (2008) Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol Ecol 17:3599–3613
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122
Neale DB, Savolainen O (2004) Association genetics of complex traits in conifers. Trends Plant Sci 9:325–330
Peakall R, Ruibal M, Lindenmayer DB (2003) Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution 57:1182–1195
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pharis RP (1966) Comparative Drought Resistance of Five Conifers and Foliage Moisture Content as a Viability Index. Ecology 47:211–221
Plummer M, Best N, Cowles K, Vines K (2006) CODA: convergence diagnosis and output analysis for MCMC. R News 6:7–11
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Prunier J, Gerardi S, Laroche J, Beaulieu J, Bousquet J (2012) Parallel and lineage-specific molecular adaptation to climate in boreal black spruce. Mol Ecol 21:4270–4286
R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R (2015) A practical guide to environmental association analysis in landscape genomics. Mol Ecol 24:4348–4370
Rousset F (2008) Genepop’007: a complete re-implementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Sambaraju KR, Carroll AL, Zhu J et al (2012) Climate change could alter the distribution of mountain pine beetle outbreaks in western Canada. Ecography 35:211–223
Sarkar NK, Kundnani P, Grover A (2013) Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 18:427–437
SAS Institute (2008) The SAS system for Windows. SAS Inst., Cary, NC, USA
Savolainen O, Pyhajarvi T, Knurr T (2007) Gene flow and local adaptation in trees. Annu Rev Ecol Evol Syst 38:595–619
Seki M, Narusaka M, Ishida J et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31:279–292
Solomon S, Qin D, Manning M et al (2007) Climate Change 2007: the physical science basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change
Steane DA, Potts BM, McLean E et al (2014) Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Mol Ecol 23:2500–2513
Taylor NL, Rudhe C, Hulett JM et al (2003) Environmental stresses inhibit and stimulate different protein import pathways in plant mitochondria. FEBS Lett 547:125–130
Thornthwaite CW (1948) An approach toward a rational classification of climate. Geogr Rev 38:55–94
Van Aken O, Zhang B, Carrie C et al (2009) Defining the mitochondrial stress response in Arabidopsis thaliana. Mol Plant 2:1310–1324
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935
Wang T, Hamann A, Spittlehouse D, Murdock TN (2012) ClimateWNA—high-resolution spatial climate data for western North America. J Appl Meteor Climatol 61:16–29
Zhu T, Jenkins MW, Lund JR (2005) Estimated impacts of climate warming on California water availability under twelve future climate scenarios. J Am Water Resour Assoc 41:1027–1038
Acknowledgments
We are very much indebted to J. Liechty for his bioinformatical assistance. We wish to express our gratitude to J. Gleason and J. Dunlap from the US Forest Service for all the logistic support when collecting seed samples at Placerville, CA, USA, and providing all the necessary metadata. A postdoctoral fellowship from the Belgian American Educational Foundation (BAEF) and a travel grant from the Research Foundation–Flanders (FWO) was granted to CV.
Data archiving statement
Genotype and environmental data have been deposited in the TreeGenes Database (accession number TGDR059).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. C. González-Martínez
Electronic supplementary material
Below is the link to the electronic supplementary material.
Esm 1
(DOCX 1748 kb)
Rights and permissions
About this article
Cite this article
Vangestel, C., Vázquez-Lobo, A., Martínez-García, P.J. et al. Patterns of neutral and adaptive genetic diversity across the natural range of sugar pine (Pinus lambertiana Dougl.). Tree Genetics & Genomes 12, 51 (2016). https://doi.org/10.1007/s11295-016-0998-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-0998-7