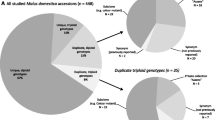
Abstract
The Research Institute of Pomology (IP), Chinese Academy of Agricultural Sciences (CAAS) in Xingcheng, China, maintains hundreds of apple accessions that originated from around the world. We have used 16 microsatellites to assess the diversity and differentiation of 391 accessions within the IP that represent Malus × domestica (from China, Japan, former Soviet Republics, and Western countries) as well as the crop’s wild relative species Malus baccata, Malus prunifolia, Malus × robusta, and Malus sieversii. We identified genetic relationships among these eight source categories that suggest that the M. × domestica cultivars from the former Soviet Republics are most closely related to M. sieversii and may represent an independent lineage of domesticated apples distinct from those found in Western Europe and North America. We show that the M. × domestica cultivars from China and Western sources are genetically similar, whereas the cultivars from Japan are distinct. We also describe two accessions of M. × domestica ssp. chinensis landraces that are believed to be over 2000 years old and are more similar to wild species than are most of the M. × domestica cultivars. We show that the wild, landrace, and cultivar accessions within the IP offer novel diversity to apple breeding programs.

Similar content being viewed by others
References
Cornille A, Giraud T, Smulders MJM, Roldán-Ruiz GP (2014) The domestication and evolutionary ecology of apples. Trends Genet 30:57–65
Cornille A, Feurtey A, Gélin U, Ropars J, Misvanderbrugge K, Gladieux P, Giraud T (2015) Anthropogenic and natural drivers of gene flow in a temperate wild fruit tree: a basis for conservation and breeding programs in apples. Evol Appl 8:373–384
Dong YC, Liu X (2006) Crops and their wild relatives in China. China Agriculture Press, Beijing, pp 55–84 (in Chinese)
Dyer RJ (2009) GeneticStudio: a suite of programs for spatial analysis of genetic-marker data. Mol Ecol Resour 9:110–113
Dyer RJ, Nason JD (2004) Population graphs: the graph-theoretic shape of genetic structure. Mol Ecol 13:1713–1728
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. doi:10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformatics Online 1:47–50
Food and Agriculture Organizations of the United Nations (2013) FAOSTAT 16 November, 2013. <http://faostat.fao.org/site/362/DesktopDefault.aspx?PageID=362>
Gardiner SE, Norelli JL, de Silva N, Fazio G, Peil A, Malnoy M, Horner M, Bowatte D, Carlisle C, Wiedow C, Wan Y, Bassett CL, Baldo AM, Celton J-M, Richter K, Aldwinckle HS, Bus VGM (2012) Putative resistance gene markers associated with quantitative trait loci for fire blight resistance in Malus ‘Robusta 5’ accessions. BMC Genet 13:25. doi:10.1186/1471-2156-13-25
Gharghani A, Zamani Z, Talaie A, Oraguzie NC, Fatahi R, Hajnajari H, Wiedow C, Gardiner SE (2009) Genetic identity and relationships of Iranian apple (Malus × domestica Borkh.) cultivars and landraces, wild Malus species and representative old apple cultivars based on simple sequence repeat (SSR) marker analysis. Genet Resour Crop Evol 56:829–842
Goudet J (1995) FSTAT, a program for IBM PC compatibles to calculate Weir and Cockerham’s (1984) estimators of F-statistics. J Hered 86:485–486
Gross BL, Henk AD, Forsline PL, Richards CM, Volk GM (2012a) Identification of interspecific hybrids among domesticated apple and its wild relatives. Tree Genet Genomes 8:1223–1235
Gross BL, Volk GM, Richards CM, Forsline PL, Fazio G, Chao CT (2012b) Identification of “duplicate” accessions within the USDA-ARS National Plant Germplasm System Malus collection. J Am Soc Hortic Sci 137:333–342
Gross BL, Volk GM, Richards CM, Reeves PA, Henk AD, Forsline PL, Szewc-McFadden A, Fazio G, Chao CT (2013) Diversity captured in the USDA-ARS National Plant Germplasm System apple core collection. J Am Soc Hortic Sci 138:375–381
Gross BL, Henk AD, Richards CM, Fazio G, Volk GM (2014) Genetic diversity in Malus × domestica (Rosaceae) through time in response to domestication. Am J Bot 101:1770–1779
Höfer M, Mohamed MA, Ali SE, Sellmann J, Peil A (2014) Phenotypic evaluation and characterization of a collection of Malus species. Genet Resour Crop Evol 61:943–964
Hokanson SC, Lamboy WF, Szewc-McFadden AK, McFerson JR (2001) Microsatellite (SSR) variation in a collection of Malus (apple) species and hybrids. Euphytica 118:281–294
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Khan MA, Olsen KM, Sovero V, Kushad MM, Korban SS (2014) Fruit quality traits have played critical roles in domestication of the apple. Plant Genome 7:1–18. doi:10.3835/plantgenome2014.04.0018
Lewis PO, Zaykin D (2001) GDA User’s manual. Department of Ecology and Evolutionary Biology. University of Connecticut. 27 July 2004. <http://lewis.eeb.uconn.edu/lewishome>
Li YN (2001) Research on germplasm resources of Malus Mill. China Agriculture Press, Beijing, pp 105–111 (in Chinese)
Liebhard R, Gianfrancechi L, Keller B, Ryder CD, Tarchini R, Van de Weg E, Gessler C (2002) Development and characterization of 140 new microsatellites in apple (Malus × domestica Borkh.). Mol Breed 10:217–241
Luby J, Forsline P, Aldwinckle H, Bus V, Geibel M (2001) Silk road apples—collection, evaluation, and utilization of Malus sieversii from Central Asia. HortSci 36:225–231
Markussen T, Kruger J, Schmidt H, Dunemann F (1995) Identification of PCR-based markers linked to the powdery-mildew-resistance gene Pl(1) from Malus robusta in cultivated apple. Plant Breed 114:530–534
Padmarasu S, Sargent DJ, Jaensch M, Kellerhals M, Tartarini S, Velasco R, Troggio M, Patocchi A (2014) Fine-mapping of the apple scab resistance locus Rvi12 (Vb) derived from ‘Hansen’s baccata #2’. Mol Breed 34:2119–2129
Perrier X, Jacquemoud-Collet JP (2006) DARwin software 2 April 2015. <http://darwin.cirad.fr/darwin>
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rambaut A (2009) FigTree v 1.3.1: Tree figure drawing tool. 4 April 2014. <http://tree.bio.ed.ac.uk/software/figtree/ >
Rambaut A, Drummond A (2009) FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh. 8 July 2015 <http://tree.bio.ed.ac.uk/software/figtree/>
Sharma YP, Pramanick KK, Sharma SK, Kashyap P (2013) Disease reaction of apple germplasm to white root rot (Dematophora necatrix). Indian J Agric Chem 70:130–134
USDA, ARS, National Genetic Resources Program (2015) Gerplasm Resources Information Network- (GRIN) [Online Database]. National Germplasm Resources Laboratory, Beltsville, MD.12 February 2015. <http://www.ars-grin.gov/cgi-bin/npgs/html/genform.pl>
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar S, Troggio M et al (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Vogt I, Wöhner T, Richter K, Flachowsky H, Sundin GW, Wensing A, Savory EA, Geider K, Day B, Hanke M-V, Peil A (2013) Gene-for-gene relationship in the host-pathogen system Malus × robusta 5-Erwinia amylovora. New Phytol 197:1262–1275
Volk GM, Henk AD, Baldo A, Fazio G, Chao CT, Richards CM (2015) Chloroplast heterogeneity and historical admixture within the genus Malus. Am J Bot (in press)
Yamamoto T, Kimura T, Sawamura Y, Manabe T, Kotobuki K, Hayashi T, Ban Y, Matsuta N (2002) Simple sequence repeats for genetic analysis in pear. Euphytica 124:129–137
Yu DJ (1979) Taxonomy of the fruit tree in China. China Agriculture Press, Beijing, pp 98–100 (in Chinese)
Zhi-Qin Z (1999) The apple genetic resources in China: the wild species and their distributions, informative characteristics and utilization. Genet Resour Crop Evol 46:599–609
Acknowledgments
We thank Professor Jia Jizheng for helpful suggestions and arrangements for the experiments performed at the Key Laboratory of Crop Germplasm and Utilization, Ministry of Agriculture, Beijing, P. R. China. This research was funded by Core Research Budget of the Non-Profit Governmental Research Institution (ICS, CAAS) and the Special Scientific Research Fund of Agricultural Public Welfare Profession of China, the Ministry of Finance, P. R. China (No. 092060302-9 and No. 201303093). This project was performed as part of the Bilateral Agreement between USDA and China’s Ministry of Science and Technology (MOST) for cooperation on US-China Agricultural Flagship Projects (Project Number 0210-22310-004-50). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Conflict of interest
The authors declare that they have no conflict of interest.
Data archiving statement
The accession and SSR data for this research are available at the Genome Database for Rosaceae (tfGDR1017; www.rosaceae.org).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Troggio
This article is part of the Topical Collection on Germplasm Diversity
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLS 66 kb)
Rights and permissions
About this article
Cite this article
Gao, Y., Liu, F., Wang, K. et al. Genetic diversity of Malus cultivars and wild relatives in the Chinese National Repository of Apple Germplasm Resources. Tree Genetics & Genomes 11, 106 (2015). https://doi.org/10.1007/s11295-015-0913-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0913-7