Abstract
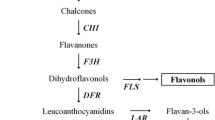
To reveal the molecular mechanisms that led to the red/green color mutation of pear between the cultivar ‘Early red Doyenne du Comice’ and its green variant strain, the full-length cDNA of the seven anthocyanin biosynthesis genes (PAL, CHS, CHI, DFR, F3H, ANS, UFGT) was cloned in both cultivars. The accession number has been submitted to National Center for Biotechnology Information (NCBI) as KC460392, KC460393, KC460394, KC460395, KC460396, KC460397, and KC460398, respectively. However, there was no sequence difference between the color mutants, which means that the skin color change was not caused by mutation of any of these genes. Meanwhile, the expression levels of these seven genes were examined by quantitative real-time PCR (qRT-PCR). Results showed that most of the structural genes were up-regulated in the red-skinned cultivar during fruit development, but the CHI and UFGT genes were highly expressed only at an early stage. The expression levels of the transcription factors MYB10, bHLH, and WD40 were also investigated by qRT-PCR, and the MYB10 gene was found to be expressed at significantly higher levels in the red variety than in the green mutant at the early stage, while the expression levels of bHLH and WD40 were higher at a later stage. These data indicate that the expression difference of structural genes in the anthocyanin biosynthesis pathway led to the skin color change of the mutant. However, MYB10, bHLH, and WD40 do not appear to be the key transcription factors that regulate the biosynthesis of anthocyanin and determine the red/green color mutant.




Similar content being viewed by others
Abbreviations
- ANS:
-
Anthocyanin synthase
- bHLH:
-
Basic helix loop helix
- CHI:
-
Chalcone isomerase
- CHS:
-
Chalcone synthase
- DAFB:
-
Days after full bloom
- DFR:
-
Dihydroflavonol-4-reductase
- F3H:
-
Flavanone 3-hydroxylase
- PAL:
-
Phenylalanine ammonialyase
- RACE:
-
Rapid amplification of cDNA ends
- qRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- SE:
-
Standard error
- TAIL-PCR:
-
Thermal asymmetric interlaced polymerase chain reaction
- UFGT:
-
UDP-glucose: flavonoid-3-O-glucosyltransferase
References
Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM (2011) Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65:771–784
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13:99–102
An XH, Tian Y, Chen KQ, Wang XF, Hao YJ (2012) The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J plant physiol 169:710–717
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48:958–970
Boss P, Davies C (2009) Molecular biology of anthocyanin accumulation in grape berries. In: Grapevine molecular physiology & biotechnology. Springer, Heidelberg, pp. 263–292
Boss PK, Davies C, Robinson SP (1996) Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol 32:565–569
Cao YF (2001) Red-skin pear germplasm of China. China Seed Industry 1, 2
Chen H, Xin G, Zhang B, Yang JY (2009) Optimization of extraction technique of anthocyanin from red peel of ‘Nanguo’ Pear. Food Science 30(8):97–100
Davies KM, Albert NW, Schwinn KE (2012) From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Functional Plant Biology 39(8):619–638
Dondini L, Pierantoni L, Ancarani V, D’Angelo M, Cho KH, Shin IS, Musacchi S, Kang SJ, Sansavini S (2008) The inheritance of the red colour character in European pear (Pyrus communis) and its map position in the mutated cultivar ‘Max Red Bartlett’. Plant Breed 127:524–526
Dussi MC, Sugar D, Wrolstad RE (1995) Characterizing and quantifying anthocyanins in red pears and the effect of light quality on fruit color. J Am Soc Hortic Sci 120:785–789
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant j cell mol biol 49:414–427
Espley RV, Brendolise C, Chagne D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP et al (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183
Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66:94–116
Feng S, Wang Y, Yang S, Xu Y, Chen X (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232:245–255
Fischer TC, Gosch C, Pfeiffer J, Halbwirth H, Halle C, Stich K, Forkmann G (2007) Flavonoid genes of pear (Pyrus communis). Trees-Struct Funct 21:521–529
Gamble J, Jaeger SR, Harker FR (2006) Preferences in pear appearance and response to novelty among Australian and New Zealand consumers. Postharvest Biol Technol 41:38–47
Gasic K, Hernandez A, Korban S (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep 22:437–438
Gonzalez A (2009) Pigment loss in response to the environment: a new role for the WD/bHLH/MYB anthocyanin regulatory complex. New Phytol 182:1–3
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780
Gu C, Wu J, Zhang SJ, Yang YN, Wu HQ, Khan MA, Zhang SL, Liu QZ (2011) Molecular analysis of eight SFB alleles and a new SFB-like gene in Prunus pseudocerasus and Prunus speciosa. Tree Genet Genome 1–12
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62:2465–2483
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585
Kim SH, Lee JR, Hong ST, Yoo YK, An G, Kim SR (2003) Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci 165:403–413
Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215:924–933
Lancaster JE, Grant JE, Lister CE, Taylor MC (1994) Skin color in apples—influence of copigmentation and plastid pigments on shade and darkness of red color in five genotypes. J Am Soc Hortic Sci 119:63–69
Niu S, Xu C, Zhang W, Zhang B, Li X, Kui L, Ferguson IB, Allan AC, Chen K (2010) Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 231:887–899
Ohno S, Hosokawa M, Hoshino A, Kitamura Y, Morita Y, Park KI, Nakashima A, Deguchi A, Tatsuzawa F, Doi M et al (2011) A bHLH transcription factor, DvIVS, is involved in regulation of anthocyanin synthesis in dahlia (Dahlia variabilis). J Exp Bot 62:5105–5116
Pierantoni L, Dondini L, Franceschi PD, Musacchi S, Winkel BSJ, Sansavini S (2010) Mapping of an anthocyanin-regulating MYB transcription factor and its expression in red and green pear, Pyrus communis. Plant Physiol Biochem 48(12):1020–1026
Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB Gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216–1232
Wei Y, Hu F, Qin Y, Wang H, Hu G (2010) Establishment and optimization of real-time PCR reaction system for gene expression in litchi pericarp (Litchi chinensis Sonn.). Chin J Trop Crop 31:7
Wu J, Zhao G, Yang Y-N, Le W-Q, Khan MA, Zhang S-L, Gu C, Huang W-J (2012a) Identification of differentially expressed genes related to coloration in red/green mutant pear (Pyrus communis L.). Tree Genetics & Genomes 9(1):75–83
Wu T, Zhang R, Gu C, Wu J, Wan H, Zhang S, Zhang S (2012b) Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. Afr J Agric Res 7:3701–3704
Zhang X, Allan AC, Yi Q, Chen L, Li K, Shu Q, Su J (2011) Differential gene expression analysis of Yunnan red pear, Pyrus pyrifolia, during fruit skin coloration. Plant Mol Biol Report 29:10
Acknowledgments
The work was financially supported by the National Public Benefit (Agricultural) Research Foundation of China (200903044), the National Science Foundation of China (30900974), and the Earmarked Fund for China Agriculture Research System (no. CARS-29).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Dandekar
Rights and permissions
About this article
Cite this article
Yang, Yn., Zhao, G., Yue, Wq. et al. Molecular cloning and gene expression differences of the anthocyanin biosynthesis-related genes in the red/green skin color mutant of pear (Pyrus communis L.). Tree Genetics & Genomes 9, 1351–1360 (2013). https://doi.org/10.1007/s11295-013-0644-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-013-0644-6