Abstract
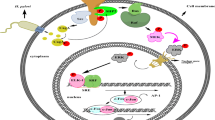
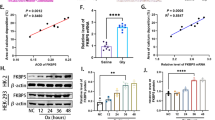
Chlamydia trachomatis is the most common bacterial pathogen causing sexually transmitted diseases. C. trachomatis infection is closely related to the development of cervical cancer, studies have shown that C. trachomatis can induce host cell autophagy. The autophagy related gene p62 plays an important role in the process of autophagy. To further understand the role of autophagy-associated gene p62 in autophagy of HeLa cells induced by C. trachomatis, p62-silencing cell line, HeLa229-shp62, and control cell line, HeLa229-shNC, were constructed, and a C. trachomatis-infected cell model was established. The autophagosome and C. trachomatis inclusions were observed under electron microscope. The autophagy level of C. trachomatis-infected HeLa cells was detected by Western blot. The results suggested that knockdown of p62 affected neither C. trachomatis infection of HeLa cells nor the initiation of C. trachomatis-induced autophagy, but at 48 h post C. trachomatis infection, autophagy levels were significantly inhibited in p62 silencing host cells. The study demonstrated the important role of p62 in the autophagy induced by C. trachomatis in HeLa cells, which could provide data support and theoretical basis for exploring the pathogenesis and prevention of C. trachomatis.



Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the article.
References
Alegre F, Moragrega AB, Polo M, Marti-Rodrigo A, Esplugues JV, Blas-Garcia A, Apostolova N (2018) Role of p62/SQSTM1 beyond autophagy: a lesson learned from drug-induced toxicity in vitro. Br J Pharmacol 175:440–455. https://doi.org/10.1111/bph.14093
Al-Younes HM, Brinkmann V, Meyer TF (2004) Interaction of Chlamydia trachomatis serovar L2 with the host autophagic pathway. Infect Immun 72:4751–4762. https://doi.org/10.1128/IAI.72.8.4751-4762.2004
Bestebroer J, V’Kovski P, Mauthe M, Reggiori F (2013) Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic 14:1029–1041. https://doi.org/10.1111/tra.12091
Cuomo F, Altucci L, Cobellis G (2019) Autophagy function and dysfunction: potential drugs as anti-cancer therapy. Cancers (Basel). https://doi.org/10.3390/cancers11101465
Lamark T, Svenning S, Johansen T (2017) Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem 61:609–624. https://doi.org/10.1042/EBC20170035
Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132:27–42. https://doi.org/10.1016/j.cell.2007.12.018
Lorincz P, Juhasz G (2019) Autophagosome-lysosome fusion. J Mol Biol. https://doi.org/10.1016/j.jmb.2019.10.028
Mackern-Oberti JP et al (2017) Male genital tract immune response against Chlamydia trachomatis infection. Reproduction 154:R99–R110. https://doi.org/10.1530/REP-16-0561
McKuen MJ, Mueller KE, Bae YS, Fields KA (2017) Fluorescence-reported allelic exchange mutagenesis reveals a role for Chlamydia trachomatis TmeA in invasion that is independent of host AHNAK. Infect Immun. https://doi.org/10.1128/IAI.00640-17
Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132. https://doi.org/10.1146/annurev-cellbio-092910-154005
Netea-Maier RT, Plantinga TS, van de Veerdonk FL, Smit JW, Netea MG (2016) Modulation of inflammation by autophagy: consequences for human disease. Autophagy 12:245–260. https://doi.org/10.1080/15548627.2015.1071759
Newman L et al (2015) Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS ONE 10:e0143304. https://doi.org/10.1371/journal.pone.0143304
O’Connell CM, Ferone ME (2016) Chlamydia trachomatis. Genital Infections. Microb Cell 3:390–403. https://doi.org/10.15698/mic2016.09.525
Paavonen J (2012) Chlamydia trachomatis infections of the female genital tract: state of the art. Ann Med 44:18–28. https://doi.org/10.3109/07853890.2010.546365
Pachikara N, Zhang H, Pan Z, Jin S, Fan H (2009) Productive Chlamydia trachomatis lymphogranuloma venereum 434 infection in cells with augmented or inactivated autophagic activities. FEMS Microbiol Lett 292:240–249. https://doi.org/10.1111/j.1574-6968.2009.01494.x
Parzych KR, Klionsky DJ (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 20:460–473. https://doi.org/10.1089/ars.2013.5371
Peipert JF (2003) Clinical practice. Genital chlamydial infections. N Engl J Med 349:2424–2430. https://doi.org/10.1056/NEJMcp030542
Ravanan P, Srikumar IF, Talwar P (2017) Autophagy: the spotlight for cellular stress responses. Life Sci 188:53–67. https://doi.org/10.1016/j.lfs.2017.08.029
Ricci V (2016) Relationship between VacA toxin and host cell autophagy in Helicobacter pylori infection of the human stomach: a few answers. Many Questions Toxins (Basel). https://doi.org/10.3390/toxins8070203
Sanchez-Martin P, Komatsu M (2018) p62/SQSTM1-steering the cell through health and disease. J Cell Sci. https://doi.org/10.1242/jcs.222836
Sharma V, Verma S, Seranova E, Sarkar S, Kumar D (2018) Selective autophagy and xenophagy in infection and disease front cell. Dev Biol 6:147. https://doi.org/10.3389/fcell.2018.00147
Subbarayal P, Karunakaran K, Winkler AC, Rother M, Gonzalez E, Meyer TF, Rudel T (2015) EphrinA2 receptor (EphA2) is an invasion and intracellular signaling receptor for Chlamydia trachomatis. PLoS Pathog 11:e1004846. https://doi.org/10.1371/journal.ppat.1004846
Tang W et al (2020) Pregnancy and fertility-related adverse outcomes associated with Chlamydia trachomatis infection: a global systematic review and meta-analysis. Sex Transm Infect 96:322–329. https://doi.org/10.1136/sextrans-2019-053999
Vergne I, Lafont F, Espert L, Esclatine A, Biard-Piechaczyk M (2017) Autophagy, ATG proteins and infectious diseases. Med Sci (Paris) 33:312–318. https://doi.org/10.1051/medsci/20173303019
Wang F et al (2019) Inflammatory mechanism of Chlamydia trachomatis-infected HeLa229cells regulated by Atg5. Biochem Biophys Res Commun. https://doi.org/10.1016/j.bbrc.2019.09.132
Witkin SS, Minis E, Athanasiou A, Leizer J, Linhares IM (2017) Chlamydia trachomatis: the persistent pathogen. Clin Vaccine Immunol. https://doi.org/10.1128/CVI.00203-17
Yang Y et al (2019) Cytoplasmic DAXX drives SQSTM1/p62 phase condensation to activate Nrf2-mediated stress response. Nat Commun 10:3759. https://doi.org/10.1038/s41467-019-11671-2
Yong EC, Chi EY, Kuo CC (1987) Differential antimicrobial activity of human mononuclear phagocytes against the human biovars of Chlamydia trachomatis. J Immunol 139:1297–1302
Yu P, Xiao L, Lin L, Tang L, Chen C, Wang F, Wang Y (2016) STAT3-mediated TLR2/4 pathway upregulation in an IFN-gamma-induced Chlamydia trachomatis persistent infection model. Pathog Dis. https://doi.org/10.1093/femspd/ftw076
Funding
This work was supported by National Natural Science Foundation of China (Grant Nos. 81371864, 81572040 and 81501791) and Natural Science Foundation of Hunan Province (2018JJ3819 and 2019JJ40392).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval
The study was approved by the ethics committee of Xiangya School of medicine, Central South University.
Consent for publication
All the authors have agreed for authorship, read and approved the manuscript, and given consent for publication of the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, F., Zhang, H., Lu, X. et al. Chlamydia trachomatis induces autophagy by p62 in HeLa cell. World J Microbiol Biotechnol 37, 50 (2021). https://doi.org/10.1007/s11274-021-03014-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-021-03014-5