Abstract
Avian polyomavirus (APV) infection, also called as budgerigar fledgling disease (BFD) causes various health problems in many psittacine species which may cause untimely death. The aims of this study were to investigate, for the first time, the detection, molecular characterization and phylogenetic analysis of avian polyomavirus (APV) in Pakistani psittacine birds. In an aviary a disease similar to APV was found and 90% of the nestlings died within a few weeks. Seven to ten-day-old parrot nestlings (n = 3) from the aviary were presented with feather abnormalities, plumage defect and were clinically depressed. Birds died at 11th, 14th and 16th day of age. Samples of hearts, livers, spleen, feathers and kidneys were collected from the dead birds. Samples were analyzed for the presence of APV DNA by using PCR. APV VP1 gene was partially sequenced, and phylogenetic analysis was performed. The APV strain was similar to those previously reported in other areas of the world. The results of this investigation indicate presence of a high frequency of APV infections in psittacine birds in Pakistan.
Similar content being viewed by others
Introduction
Budgerigar fledgling disease (BFD) is a contagious disease causing high mortality in fledgling budgerigars (Melopsittacus undulatus), ring-necks parrots (Psittacula krameri), lovebirds (Agapornis spp.), conures (Aratinga nenday, Pyrrhura spp.) and macaws (Ara spp.) (Kingston 1992; Piasecki and Wieliczko 2010). However, the disease has also been described in Passeriformes and Falconiformes (Johne and Müller 1998). It was first reported in the United States and Canada and subsequently has been recognized in other countries (Bernier et al. 1981; Davis et al. 1981). BFDV is a member of the family Polyomaviridae; it was placed in the genus Polyomavirus in the family Papovaviridae previously. Thus BFDV is generally called avian polyomavirus (APV) (Ogawa et al. 2005). BFDV is a non-enveloped icosahedral DNA virus with a diameter of 40–50 nm. The genome of BFDV is double-stranded circular DNA and the size is about 4,980 bp. It has been characterized as the first non-mammalian member of the polyomavirus genus (Rott et al. 1988). APV infection causes acute death, abdominal distention and abnormal feathering (French molt) on the back and abdomen, lack of filoplumes on the head and neck, and hemorrhage on subcutaneous and subserosal regions. Pathologic examinations show hydropericardium, heart and liver enlargement, congested kidneys, hemorrhage within the body cavities and immune complex glomerulopathy (Katoh et al. 2009). Large basophilic nuclear inclusion bodies in spleen, liver, and kidneys are found on histopathological examinations. In budgerigars APV infection is often subclinical and birds die without showing any signs and symptoms (Katoh et al. 2010). This report describes the detection and molecular characterization of APV in a small aviary in Pakistan for the first time and briefly reviews APV infection in pet birds.
Material and methods
Three parrots were presented for diagnostic evaluation during February to April, 2016 in the Clinical facility of Faculty of Veterinary and Animal Sciences PMAS-Arid Agriculture University Rawalpindi. The age of birds at the time of presentation was less than 10 days. The birds were kept under observation. Death of birds occurred at 11, 14 and 16th days of age. Owner of the birds reported that young birds in the aviary most often died between 10 and 21 days of age and those that survived were stunted and lacked normal feather development.
Samples collection
Samples of hearts, livers, spleens, feathers and kidneys were collected from the dead birds. Each sample was homogenized separately in phosphate buffered saline solution. Samples were subjected to freeze and thaw cycle for three times, centrifuged at 12,000 rpm for 10 min and supernatant was separated and stored at -20 °C.
DNA isolation
DNA isolation from the hearts, livers, spleens, feathers and kidneys was performed. After the dissection of dead birds and the preparation of organs, DNA was isolated from the respective tissue using the PureLink® Genomic DNA Kit (Invitrogen). DNA concentration in the samples was measured using Nano-Drop spectrophotometer. DNA samples were stored at -20 °C.
PCR amplification of viral DNA
Primers (Macrogen, Korea) were designed for a fragment of VP1 gene sequence (APV genome position 2182–2207 to 2733–2710). The primer sequences for forward primer was 5′-CTTATGTGGGAGGCTGCAGTGTT-3′ and reverse primer was 5′-TACTGAATAGCGTGGTAGGCCTC-3′ (Katoh et al. 2009).
To perform PCR, Taq 2X Master Mix (NEB, UK) was used. Approximately 100 ng/μl the extracted DNA was used in a total volume of 50 μl reaction mixture for PCR. The thermocycler conditions were; Initial denaturation for 4 min at 94 °C, followed by 40 cycles of 30s at 94 °C, 1 min at 51 °C and 1 min at 72 °C and final extension at for 7 min at 72 °C. For negative control nuclease free water was used. A PCR product of 550 bp, was separated on 1.5% agarose gels.
Sequence and phylogenetic analysis
One of the positive PCR Products from each bird were further subjected to sequencing and phylogenetic analyses. For gene sequencing, the samples were sent to Macrogen® Korea using ABI 3730xL, standard DNA sequencer.
Sequences derived from this study and those obtained from the GenBank database were aligned by the CLUSTAL_W method in the software Seaview®. The distances were computed mean-wise and overall using MEGA7®.The gene sequences were translated using Seaview®. Sequences were subsequently analyzed with neighbor joining to construct the phylogenetic tree (Kumar et al. 2016). The statistical significance of the relationships obtained was determined by bootstrap re-sampling analysis wit 1000 repetitions. The sequences were deposited to GenBank database. The accession numbers were assigned by NCBI for APV VPI gene sequences from the present study; MG946129, MG946130 and MG946131 and for their amino acid sequences were AVM41561, AVM41562 and AVM41563. A comparison was made among the sequences of APV from this study with APV sequences of other studies (KT203766.1, GU452537.1, AB453165, FJ385773, AF241170, KT203768, KM092491, DQ304719 and DQ074760) (Table 1).
Results
Three young parrots were presented for diagnostic evaluation. Parrots died at the age of 11, 14, 16 days. Aviary owner reported that death rate in young parrots had markedly increased from 10% to 90% in 6 months. Most of the young birds died between 10 and 21 days of age.
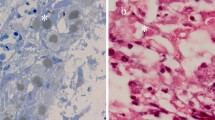
The presented birds were small as compared to unaffected nestlings of same age. They lacked feathers on the head, neck, back and breast regions and had underdeveloped flight feathers and tail feathers. They crops were full and abdomens were distended, with undigested food. Hemorrhages were prominent on neck, chest, shoulder and thigh muscles as well as on liver, heart, bursa and intestines. Livers were red mottled (Fig. 1). Postmortem lesions were similar in all birds including ascites, hydropericardium and hepatomegaly with hemorrhagic mottling. (Fig. 1). The diagnosis of APV was made on the basis of gross lesions and molecular diagnosis was made on the basis of PCR.
PCR detection of the major capsid protein VP1 gene
DNA was extracted from heart, liver, spleen, kidney and feathers of each bird separately. PCR was performed on the specific primer of the VP1 gene was used to test APV. A band of 550 bp of the specific amplified fragment was obtained for each of the 15 tissue samples (five tissues from each bird) (Fig. 2). It was shown that the all the birds were infected with APV.
PCR products of DNA from tissues of three APV positive birds (a and b). Heart (H), Liver (L), Spleen (S), Kidney (K), and Feathers (F) were used from each bird for DNA isolation. A PCR product of 550-bp was considered positive. For electrophoresis 1.5% gel was used. Lane M: 100-bp DNA ladder, Lane N: Negative control
The positive PCR products were purified and one PCR product from each bird was sent for DNA sequencing. The results of the sequence analysis showed that all 3 sequences were of APV-VP1 genes, and the similarities among them were 99%–100%. The three nucleotide sequences were deposited in GenBank and the assigned accession numbers were MG946129, MG946130 and MG946131 (For names see Table 1). Based on BLAST (NCBI), the similarity of the VP1 gene among these and the other isolates was more than 95% which suggests that the VP1 gene was conservative for APV (Fig. 3).
Phylogenetic analysis using maximum likelihood method of three DNA sequences (of 550-bp region each) of VPI gene of APV. (The Accesssion Numbers MG946129, MG946130 and MG946131 were designated by NCBI to the DNA sequences from the present study and a phylogenetic tree was constructed based on alignment with highly similar sequences of VP1 genes retrieved from NCBI database that showed high homology with our sequences. Evolutionary analyses were conducted in MEGA7 software
Molecular phylogenetic analysis of major capsid protein VP1 gene region of APV
The sequences of the major capsid protein VP1 gene region of 3 APV-positive samples were compared with 9 sequences reported by other researchers (Table 1). A phylogenetic tree was constructed based on alignment with those sequences retrieved from NCBI database that showed high homology with our sequences. The phylogenetic analysis demonstrated that the sequence of the VP1 gene obtained in the present study were showing 99 to 100% homology with most of the Polyomavirus VP1 gene sequences worldwide. There are two branches in the tree with high bootstrap values. APV strains isolated from Poland (KT203766.1), China (GU452537.1 and FJ385773.1), Japan (AB453165.1), Brazil (KM092491.1), New Zealand (KT203768.1), Germany (AF241170.1) and Taiwan (DQ304719.1) formed one complex branch whereas sequence from Czech Republic (DQ074760.1) formed a single branch. The big branch can be separated into two smaller branches: KT203766.1, GU452537.1 and AB453165.1 belonged to one branch, and KM092491.1, KT203768.1, AF241170.1, FJ385773.1 and DQ304719.1 in other branch.
Discussion
The Avian Polyomavirus is a highly infectious psittacine virus, reaching almost 100% infection rates in indoor aviaries (Philadelpho et al. 2015). The disease has been described in USA (Dolz et al. 2013; Varsani et al. 2015), Europe (Johne et al. 2006), South Africa (Tajbhai et al. 2004), Asia (Ogawa et al. 2006; Zhuang et al. 2012), New Zealand (Tajbhai et al. 2004) and Australia (Hulbert et al. 2015).
To the best of our knowledge this is the first report of presence of APV in young parrots in Pakistan. Three birds of age ranges between 7 and 16 days were presented to the clinical facility. Initially the birds appeared dull and anorexic, abnormal feathers, skin discoloration, and abdominal distension were the most common clinical observations. Clinical signs for APV in young parrots are variable and mostly depend on the species and the age and deaths have been reported without any premonitory signs of the disease in budgrigars (Altan et al. 2016). In the present study the birds died within 7 days of presentation. Gross lesion and haemorrhages on the shoulder, chest, thigh muscels, liver and heart were indicative of APV infection. Mostly clinical signs due to APV infection are not specific and are present in several differential diagnoses, it is often neglected by the veterinarian (Ogawa et al. 2005). However, molecular diagnosis using PCR can confirm the diagnosis. Another virus psittacine beak and feather disease (PBFDV) and APV infections cause similar feather disorders and cannot be differentiated by clinical diagnosis. PCR is used as a highly sensitive method, capable of detecting specific DNA sequences in various biological samples and has been used extensively in the diagnosis as well as confirmation of APV and PBFDV infection (Altan et al. 2016; Dolz et al. 2013; Ogawa et al. 2005). Ogawa et al. (2005) used duplex shuttle PCR for the differential diagnosis of APV and PBFDV in psittacine birds. In the present study PCR results confirmed the presence of APV-DNA in heart, spleen, kidney, liver and feathers from each the birds (Fig. 2). PCR was performed for the segment of APV major capsid VP1 Protein gene which is a conserved region in APVs. Three positive PCR products (one from each bird) showing a 550-bp band on gel were sent for DNA sequencing. Results further confirmed the presence of APV in the samples. Sequences were submitted to NCBI Genbank and accession numbers were assigned to those (MG946129, MG946130 and MG946131). The three of the sequences were 100% identical showing that all the birds were infected with similar strain of the APV.
Phylogenetic analysis have shown that APV has no geographic separation. As shown in the phylogenetic tree, the geographic distribution of hosts in the big branch included Asia, North America, Europe, and Australia, which suggests that APV has a worldwide distribution. The transmission route might include bird migrations, free trading of poultry, dirty bird transportation, and so on (Kou et al. 2008; Reavill and Dorrestein 2018). These findings indicate that further molecular and epidemiologic investigation of APV is needed to prevent its transmission and to protect the breeding industry in psittacine birds.
APV also infect some kind of poultry besides all types of parrots. Although APV does not induce death in poultry but the APV infection in poultry should be monitored, and surveillance of poultry is both important and necessary (Katoh et al. 2010). This article is the first report on the presence of APV in Pakistan. The DNA sequencing and phylogenetic analysis revealed that the virus has the same genotype as those in other regions. Poultry sector of Pakistan is rapidly growing industry and important measures should be taken to keep the poultry free from different infections like APV. This study will be a useful in further diagnoses of APV in other birds in Pakistan and in vaccine development against APV.
References
Altan E, Eravci E, Cizmecigil UY, Yildar E, Aydin O, Turan N, Ozsoy S, Tekelioglu KB, Kurt T, Ozsemir KG (2016) Detection and phylogeny of beak and feather disease virus and avian polyomavirus in psittacine pet birds in Turkey. J Exot Pet Med 25:280–287
Bernier G, Morin M, Marsolais G (1981) A generalized inclusion body disease in the budgerigar (Melopsittacus undulatus) caused by a papovavirus-like agent. Avian Dis 25:1083–1092
Davis R, Bozeman L, Gaudry D, Fletcher O, Lukert P, Dykstra M (1981) A viral disease of fledgling budgerigars. Avian Dis 25(1):179–183
Dolz G, Sheleby-Elías J, Romero-Zuñiga JJ, Vargas-Leitón B, Gutiérrez-Espeleta G, Madriz-Ordeñana K (2013) Prevalence of psittacine beak and feather disease virus and avian polyomavirus in captivity psittacines from Costa Rica. Open J Vet Med 3:240–245
Hulbert CL, Chamings A, Hewson K, Steer P, Gosbell M, Noormohammadi A (2015) Survey of captive parrot populations around Port Phillip Bay, Victoria, Australia, for psittacine beak and feather disease virus, avian polyomavirus and psittacine adenovirus. Aust Vet J 93:287–292
Johne R, Müller H (1998) Avian polyomavirus in wild birds: genome analysis of isolates from Falconiformes and Psittaciformes. Arch Virol 143:1501–1512
Johne R, Wittig W, Fernández-de-Luco D, Höfle U, Müller H (2006) Characterization of two novel polyomaviruses of birds by using multiply primed rolling-circle amplification of their genomes. J Virol 80:3523–3531
Katoh H, Ohya K, Une Y, Yamaguchi T, Fukushi H (2009) Molecular characterization of avian polyomavirus isolated from psittacine birds based on the whole genome sequence analysis. Vet Microbiol 138:69–77
Katoh H, Ogawa H, Ohya K, Fukushi H (2010) A review of DNA viral infections in psittacine birds. J Vet Med Sci 72:1099–1106
Kingston RS (1992) Budgerigar fledgling disease (papovavirus) in pet birds. J Vet Diagn Investig 4:455–458
Kou Z, Zhang Z, Chen S, Fan Z, Tang S, Zhao L, Li T (2008) Molecular characterizations of avian polyomavirus isolated from budgerigar in China. Avian Dis 52:451–454
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Ogawa H, Yamaguchi T, Fukushi H (2005) Duplex shuttle PCR for differential diagnosis of budgerigar fledgling disease and psittacine beak and feather disease. Microbiol Immunol 49:227–237
Ogawa H, Chahota R, Hagino T, Ohya K, Yamaguchi T, Fukushi H (2006) A survey of avian polyomavirus (APV) infection in imported and domestic bred psittacine birds in Japan. J Vet Med Sci 68:743–745
Philadelpho NA, Guimarães MB, Piantino Ferreira AJ (2015) A case report of Avian polyomavirus infection in a blue fronted parrot (Amazona aestiva) associated with anemia. Case Reports in Veterinary Medicine 2015
Piasecki T, Wieliczko A (2010) Detection of beak and feather disease virus and avian polyomavirus DNA in psittacine birds in Poland. Bull Vet Inst Pulawy 54:141–146
Reavill DR, Dorrestein G (2018) Psittacines, Coliiformes, Musophagiformes, Cuculiformes. In: Pathology of Wildlife and Zoo Animals. Elsevier Academic Press, New York, pp 775–798
Rott O, Kröger M, Müller H, Hobom G (1988) The genome of budgerigar fledgling disease virus, an avian polyomavirus. Virology 165:74–86
Tajbhai K, Bragg R, Albertyn J (2004) Psittacine beak and feather disease virus in budgerigars and ring-neck parakeets in South Africa. Onderstepoort J Vet Res 71:29–34
Varsani A, Porzig EL, Jennings S, Kraberger S, Farkas K, Julian L, Massaro M, Ballard G, Ainley DG (2015) Identification of an avian polyomavirus associated with Adelie penguins (Pygoscelis adeliae). J Gen Virol 96:851–857
Zhuang Q, Chen J, Mushtaq MH, Chen J, Liu S, Hou G, Li J, Huang B, Jiang W (2012) Prevalence and genetic characterization of avian polyomavirus and psittacine beak and feather disease virus isolated from budgerigars in mainland China. Arch Virol 157:53–61
Acknowledgements
We would like to thank Dr. Inga Dry of Roslin Institute, University of Edinburgh UK, for her technical support in experiment and manuscript preparation.
Funding
This work was funded by Pir Mahr Ali Shah, Arid Agriculture University Rawalpindi, Pakistan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Riaz, A., Yousaf, A., Moaeen-ud-Din, M. et al. First detection and molecular characterization of avian polyomavirus in young parrots in Pakistan. Vet Res Commun 43, 197–202 (2019). https://doi.org/10.1007/s11259-019-09759-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11259-019-09759-7