Abstract
Although cowpea is an important food security crop in Africa, its production is constrained by insect pests such as the legume pod borer (Maruca vitrata) (Lepidoptera). Potential control strategies have focussed on using insecticidal toxins such as the crystal Cry proteins and vegetative insecticidal proteins (Vips) encoded by the cry and vip genes, respectively, of the bacterium Bacillus thuringiensis (Bt). This study sought to identify vip genes encoding toxins active against Maruca Pod Borer (MPB), from Australian Bt isolates. A collection of 224 Bt isolates was screened with gene-specific primers to identify those containing target vip genes namely vip3Aa35, vip3Af1, vip3Ag, vip3Ca2 and vip3Ba1. The coding sequences of the vip3 genes were cloned and over-expressed in Escherichia coli to produce Vip3 protein. The proteins were incorporated into Maruca artificial diets for use in insect bioassays with MPB larvae to screen for toxicity. Of these, Vip3Ba1 protein was found to strongly inhibit larval growth and was selected as the candidate gene for cowpea transformation. A vip3Ba gene reconstructed for plant expression was used in transforming cowpea via Agrobacterium tumefaciens. Transgenic lines expressing Vip3Ba protein were used in insect feeding trials to assess protection against Maruca and were found to be completely protected from this pest. We propose that the vip-cowpea lines could be combined with existing cry-transgenic cowpea to introgress this additional resistance trait and thus avoid or greatly delay the development of resistance in Maruca.
Similar content being viewed by others
Introduction
Cowpea is an important grain legume in the developing world (Timko and Singh 2008). It is cultivated in the tropics with West Africa producing 4.5 million tonnes of dry grain in 2014 alone (FAOSTAT 2016). Nigeria is the largest cowpea-producing country in Africa (Jacob et al. 2016). Cowpea is adapted to the savannah region because of its drought tolerance (Boukar et al. 2013) and is grown by resource-poor farmers for multiple uses including food and fodder (Murdock et al. 2008; Nkongolo et al. 2009). Cowpea production is constrained by diseases and pests, with insects causing significant economic losses. The insect pests that attack cowpea include pod sucking bugs, weevils, flower bud thrips and pod borers (Singh and van Emden 1979), and are responsible for significant cowpea losses (Jackai and Daoust 1986; Jackai 1995). The Maruca pod borer (MPB), Maruca vitrata (Lepidoptera; Crambidae), also known as the legume pod borer, is a pest that causes large losses in cowpea yields (Singh and Van Emden 1979; Jackai 1995; Tamo et al. 2003). MPB is found within the tropical and sub-tropical regions, but is most devastating in sub-Saharan Africa (Margam et al. 2011). This insect pest is responsible for losses of up to 80% if no control measures are employed. The larva is the most destructive stage of this pest and feeds on flower parts, green pods and seeds of cowpea and several other leguminous crops (Singh and Van Emden 1979; Jackai and Daoust 1986; Mohammed et al. 2014). Although MPB infestation has been controlled using insecticides and microbial biopesticide formulations (Taylor 1968), farmers in developing countries do not spray as they can rarely afford the chemicals (Harwood 1979). Furthermore, excessive use of insecticides can be detrimental to human health and the environment if the correct dosage is not used (Pimentel et al. 1992). The development of host plant resistance has been very successful in controlling certain insect pests in cowpea. Lines with resistance to the cowpea curculio beetle, aphids and flower thrips have been developed in national breeding programmes and adopted in several countries (Hall et al. 2003). Breeding for host plant resistance, however, has not proved to be practical for MPB. Although wild Vigna species containing genes for resistance to MPB exist, these genes cannot be crossed into cowpea due to sexual incompatibility (Fatokun 2002). As such, genetic engineering has been proposed as an option to improve cowpea yields (Machuka et al. 2000). One control strategy for MPB is based on the deployment of insect-resistant transgenic crops producing highly specific insecticidal proteins (cry toxins) from the soil bacterium Bacillus thuringiensis (Bt), so called Bt crops. Genes conferring resistance against other pests have been isolated from Bt and introduced into crops, such as corn, cotton, chickpea and pigeon pea and are effectively protected against targeted insect pests (Bravo et al. 2005; Chakraborty et al. 2016; Kaur et al. 2016) resulting in increased yields and reduced insecticide applications (Qaim 2009). Recently, a cry transgene has been used to generate many cowpea lines expressing Cry 1Ab protein and one line was selected as a breeding parent following several years of field trialling against Maruca in the field in West Africa (Higgins et al. 2012; Mohammed et al. 2014).
Although Bt crops have been very successful, studies have shown that insects can develop field-evolved resistance to specific Bt crops, particularly in those carrying a single Bt gene (Wilson et al. 1992). As such, it is possible that Maruca could eventually develop resistance to the Cry 1Ab protein. Stacking two resistance genes with differing mechanisms of action can substantially reduce the likelihood of development of resistance (Roush 1998). One such resistance gene class is the vip gene(s) of Bt, which encode the vegetative insecticidal proteins (Vips). The Vips are synthesized during the vegetative growth phase of the bacterium (Estruch et al. 1996), bear no amino acid sequence similarity to Cry proteins and have a different mechanism of action (Lee et al. 2003; Gouffon et al. 2011). Hence, they have been used to complement Cry proteins for insect resistance management in cotton (Whitehouse et al. 2007). Vip toxins with insecticidal activity are grouped according to their specificity for target insects. The Vips have been shown to be active against a range of insect pests. The Vip1 and Vip2 toxins are binary proteins active against coleopteran (Warren 1997) and homopteran pests (Yu et al. 2011a), respectively. Vip3 toxins are active against lepidopteran insects (Estruch et al. 1996) making them potential candidates for MPB management. Vips are activated by insect gut proteases upon ingestion and subsequently interact with specific receptors found in the midgut epithelial tissue. This leads to formation of pores, rupture of the epithelial cells and eventual cell death (Lee et al. 2003). There are more than 70 vip genes encoding different Vip3 proteins listed in the Crickmore database (http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/). These genes are classified into three families, namely 3A, 3B and 3C, and further grouped into nine sub-families in 3A (Vip3Aa to Ai), two 3B sub-families (Vip3Ba and Bb) and one 3C family member (Vip3Ca) based on nucleotide sequence similarities (Crickmore et al. 2014).
This study aimed at identifying a vip3 gene product with activity against MPB and therefore with potential to improve insect resistance management in cowpea. Representatives of five vip3 gene groups were isolated, cloned and expressed in Escherichia coli (E. coli) to produce Vip3 protein for insect bioassays. A candidate gene encoding a Vip3Ba toxin was selected as a potential source for MPB management in cowpea and subsequently reconstructed for plant expression and used in cowpea transformation. Transgenic lines expressing the vip3Ba gene were fully protected against MPB larvae in insect feeding studies.
Materials and methods
Screening Bt strains for the presence of vip3 genes
The primer pairs were designed using DNA sequences of vip3 genes obtained from the National Centre for Biotechnology Information (NCBI) database. To identify vip3 family-specific genes, sequences were subjected to a multiple sequence alignment in FASTA format using the Clustal W version 2.0 program hosted at the European Bioinformatics Institute (EBI) (Larkin et al. 2007) with default settings to identify conserved regions for primer designing (Table S1 in Supplementary material 1; PCR Set A). To design primers for specific vip3 gene fragments (Table S1 in Supplementary material 1; PCR Set B) and to amplify the entire coding regions (Table S1 in Supplementary material 1; PCR Set C), the target gene sequences were retrieved from the NCBI database. In order for the primers in Set C to be compatible with the expression vector, a His tag sequence (18 nucleotides long) was added to each ORF after the start codon for protein purification (Table S1 in Supplementary material 1; PCR Set C).
Cloning of vip3 and cry2Aa genes in E. coli
The full coding regions of vip3 genes, vip3Aa35, vip3Ag and vip3Ba1, were isolated from Bt DNA by PCR amplification while the full ORFs of vip3Af1 and vip3Ca2 were synthesized by GeneArt synthesis, Life Technologies Australia based on accession numbers AJ872070 and JF916462, respectively. These were inserted into a pMk-RQ (kanR) vector flanked by a kanamycin resistance gene and an origin of replication site, Col E1 (Life Technologies, Australia), and were subsequently amplified and isolated from the plasmids. The PCR products of all five genes were purified and transferred into pETite vectors (Lucigen Corporation, 2905 Parmenter St, Middleton, WI 53562, USA). To confirm the presence of the insert in the expression vector, plasmid DNA was extracted and either used as a template for PCR analysis or for restriction mapping and finally, DNA sequencing. The plasmid DNA of the positive clones was used to transform E. coli strain BL21 (DE3) for protein expression. The cry2Aa gene was previously cloned by Dan Pikler (Personal Communication) and was expressed in E. coli strain BL21 and used here as a positive control.
Protein expression and analysis
Escherichia coli transformed with the vip3 genes or cry2Aa were grown overnight at 37 °C and protein was extracted from the cultures as described by Studier (2005). The protein concentrations of the supernatant and pellet fractions were determined according to Bradford (1976) and 30 µg of protein was loaded on 10% Bis-Tris precast NuPage gels (Invitrogen) for SDS–PAGE analysis. Untransformed E. coli (BL21) cells were used as a negative control and E. coli transformed with cry2Aa was used as a positive control (Srinivasan 2008). Mass spectrometry of selected protein bands was carried out at the Australian Proteome Analysis Facility (Macquarie University, Sydney, NSW 2109, Australia) to identify the expressed Vip3 proteins. The resulting data was generated using Analyst 2.0 MASCOT script searched by Mascot and subsequently subjected to database matching (Cottrell 2011). The protein identification program used for the analysis assigns a score for peptides that match the predicted fragments based on the peptide sequences in the database (Matrix science 2014, http://www.matrixscience.com/).
Insect bioassays using E coli-expressed protein
A Maruca colony was maintained as described by Jackai and Raulston (1988). The insects were reared in a growth room set at 25 °C and 50% relative humidity (RH) for optimal growth and mating conditions. A replicated series of bioassays using MPB larvae was carried out with proteins encoded by the gene representing each of the five Vip groups. Five concentrations of each Vip3 protein (partially purified by using fractions enriched for the Vip protein) were used in preparing artificial MPB diets: 0, 3, 10, 30 and 100 µg of Vip3 protein per gram of diet. Diets with Cry2Aa protein and without Bt protein represented positive and negative controls, respectively. To calculate the amount of Vip or Cry protein to be incorporated in the artificial diet, the amount of Vip or Cry toxin present was estimated based on the Vip protein bands as seen on the Coomassie stained gels (Fig. S1). The bacterial proteins were dissolved in 50 mM sodium carbonate (pH 11.2) and incorporated into 50 ml of diet. For Vip3Ba protein, both the soluble and insoluble fractions were used. The diet was dispensed into Petri dishes and allowed to solidify at room temperature. Prior to commencing the assays, the diet was further divided into 16 cubes of ~2.5 g each, and placed in 16 individual wells of a plastic feeding tray. For each protein concentration, one first-instar larva was introduced into each well containing one cube of diet (i.e., N = 16 larvae per concentration) and sealed with a perforated lid using a hot sealer. These were incubated in a climate chamber at 25 °C and 50% RH. After 10 days, when larvae in the negative control experiments reached their fifth instar, mortality was scored and the weight of all surviving larvae was recorded using a digital microbalance. There were three separate bioassays conducted for each protein. Mortality and average weight of surviving larvae was calculated from the pooled data of all bioassays. The data were analysed using Analysis of Variance (ANOVA).
Construction of a Vip3Ba gene for plant expression
To enhance the expression of the Bt vip3Ba gene in cowpea, the coding region was modified according to Perlak et al. (1991). The GC content was increased, polyadenylation signals and mRNA destabilizing sequence motifs were deleted and plant-preferred codons were used. The re-designed gene was synthesized by GeneArt synthesis, Life Technologies Australia Pvt Ltd (Mulgrave VIC 3170 Australia) and inserted into an Agrobacterium tumefaciens- binary vector based on pART27 (Gleave 1992) but in which the Nos-NptII was replaced with an S1-Npt II from pPLEX 502 (Schunmann et al. 2003) using the assembly method described by Gibson (2011). The expression of the vip3Ba gene was under the control of the Arabidopsis small subunit 1A promoter and Nicotiana tabacum small subunit 3′ end derived from pSF12 (Tabe et al. 1995). Electro-competent A. tumefaciens AGL1 cells were transformed with the binary vector by electroporation.
Cowpea transformation
The reconstructed vip3Ba gene was used to transform cowpea based on a modified version of the protocol by Popelka et al. (2006). In brief, cowpea seeds were prepared for Agrobacterium infection, which included a sonication step, followed by a 3-day co-cultivation step. For regeneration, the explants were subcultured on shoot induction and shoot elongation media containing Murashige and Skoog (MS) salts (Murashige and Skoog 1962). They were subjected to a stringent selection regime of kanamycin at increasing concentrations (100 and 150 mg/L kanamycin) and intermittent geneticin (30 mg/L), until the rooting stage. Molecular characterization by PCR and western blotting of the putative independent transgenic lines was carried out to confirm the presence and expression of the transgene, and the transgenic lines were subsequently acclimatized and transferred to the glasshouse.
Insect bioassays using transgenic leaf material
Leaves of transgenic cowpea lines expressing Vip3Ba were fed to Maruca larvae. Bt cowpea line 709A expressing Cry1Ab (Higgins et al. 2012) was used as a positive control while the non-transgenic parent line IT86D was used as a negative control. Feeding trays consisted of 32 wells (1 cm diameter × 1 cm height). For each line, 16 wells were partially filled with 5 mL of 1% water agar containing 0.1% (w/v) sorbic acid to avoid fungal contamination (Acharjee et al. 2010). Approximately 20 mg of fresh leaf material was placed on the agar in each well.
One 6-day old larva (2.2 ± 0.1 mg) was introduced onto each leaf and the trays were sealed with a perforated lid using a hot sealer. The assays were done in a climate chamber at 25 °C and 50% RH. Surviving larvae were re-fed with fresh leaf every other day. After 10 days, the bioassay results were scored; mortality and final weights of surviving larvae were recorded. Three separate bioassay experiments were conducted for each cowpea line, resulting in a minimum of 48 replications per treatment.
Results
Identification, cloning and the expression of vip3 genes in E. coli
A number of primer pairs were used to screen an Australian collection of Bt for the presence of vip3 genes (Beard et al. 2008) and to enable the amplification of selected open reading frames (ORFs) for subsequent protein expression in E. coli. A bioinformatics approach was initially used to identify and group a diverse range of Vip3 proteins (Crickmore et al. 2014) as candidates for expression in E. coli and testing for insecticidal activity against MPB. The amino acid sequences of the proteins encoded by vip3 (3A, 3B and 3C) genes were compared. The majority (93/102) of the vip3 genes in the database belong to the 3A family with 61 of the 3A sequences belonging to the “Aa” subfamily. Analysis of these 61 sequences using a pairwise sequence alignment tool (EMBOSS Water) (http://www.ebi.ac.uk) revealed they were more than 95% identical. Based on this analysis, protein Vip3Aa35 was arbitrarily selected as the point of reference and was designated to represent Group 1. Alignment of the amino acid sequence of Vip3Aa35 with all other Vip3 proteins revealed that the other Vip3 proteins clustered into four groups (designated 2–5) with between 90 and 60%, identity with Vip3Aa35, respectively (Table 1). Based on these sequence comparisons, the proteins Vip3Aa35 (Group 1), Vip3Af1 (Group 2), Vip3Ag (Group 3), Vip3Ca2 (Group 4) and Vip3Ba1 (Group 5) were selected as the candidate proteins for further analysis. Following the identification of Bt strains containing the selected vip3A and vip3B genes (data not shown), the DNA of the selected strains was subsequently used as a template to amplify the full ORFs of the genes for cloning into an E. coli protein expression vector. No Bt strains containing a vip3C gene were identified and no amplification was observed for vip3Af1 gene. Therefore, the coding sequences of vip3Af and vip3Ca2 were chemically synthesized.
The plasmids containing the five vip3 genes (vip3Aa35, vip3Af1, vip3Ag, vip3Ba1 and vip3Ca2) were transformed into E. coli strain BL21 for protein expression. As a positive control, cry2Aa was also expressed in E. coli. Soluble and insoluble protein fractions were extracted from each of the five vip3-transformed cultures and the cry2Aa-transformed culture and these were analysed for the presence of the Vip and Cry2Aa proteins using SDS–PAGE and mass spectrometry (MS). Equivalent fractions from untransformed E. coli were also extracted as controls. When the extracts were analysed by SDS–PAGE, major bands migrating at Mr 75,000 were present in extracts of E. coli transformed with all five genes (Fig. S1 in Supplementary material 2). Unique high molecular weight bands (estimated to be >250 kDa) were also observed in the soluble fractions. When extracts from Cry2Aa-transformed and non-transformed E. coli were analysed, a major band of the size expected for the Cry2Aa protein (~65 kDa) was present in the insoluble fraction from the Cry2Aa-transformed cells (Fig. S1 in Supplementary material 2). In addition to the ~65 kDa band, several additional lower MW bands were also present in the insoluble fraction. To determine whether the protein bands migrating at Mr 75,000 derived from the five vip3-transformed cultures were the target Vips, the bands were excised from the SDS–PAGE gels and analysed by MS/MS. Subsequent interpretation of the MS/MS data using the Mascot database revealed that the highest Mascot scores were with Vip sequences. The protein coverage of Vip3A proteins ranged between 55 and 70%, Vip3Ca was 70% and Vip3Ba protein coverage was 76%. Based on this data, it was considered highly likely that the Mr 75,000 bands were indeed the target Vips (Table 1). When one of the high molecular weight bands (>250 kDa) from the soluble fraction from Group 2 was subjected to mass spectrometry analysis it was found to be composed of Vip3Af1 sequences with a Mascot score of 1347 and 39% coverage (data not shown).
Vip3Ba protein inhibited the growth of Maruca larvae
A replicated series of bioassays using MPB larvae was carried out to test the insecticidal activity of the bacterially-expressed Vip3Aa35, Vip3Af1, Vip3Ag, Vip3Ba1, Vip3Ca2 and Cry2Aa proteins. Five concentrations (0, 3, 10, 30 and 100 µg/g diet) of each freeze-dried, partially purified Vip3 protein and Cry2Aa protein were incorporated into an artificial MPB diet and the effect on MPB larvae was investigated after a 10-day growth period. The average weight of negative control larvae fed on diets without Vips or Cry2Aa protein consistently ranged between 70 and 80 mg while the average weight of positive control larvae was 6 mg at 3 µg/g of diet after 10 days. Further, all larvae fed on the negative control diet developed to the fifth-instar while mortality was observed on larvae fed on positive control at 10 µg/g of diet. On average, the growth of surviving larvae was reduced by over 90% at 3 µg Cry2Aa per g diet (Fig. 1) and they only developed to second instars. The diets containing Vip3Aa35, Vip3Af1, Vip3Ag and Vip3Ca2 had little or no effect on the growth or development of MPB larvae and all larvae developed to fifth instars (Fig. 1). In contrast, the growth of larvae fed on a diet containing Vip3Ba1 was reduced by 83, 84, 72 and 89% at concentrations of 3, 10, 30 and 100 µg/g of diet, respectively (Fig. 1; Table S2 in Supplementary material 3). Most surviving larvae on the Vip3Ba diet only developed to third instars.
Reconstruction of the vip3Ba gene for cowpea transformation
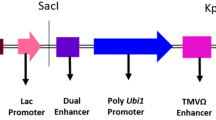
Prior to transformation into cowpea, the ORF of the vip3Ba gene was optimized for plant expression by increasing the GC content from approximately 30–50%, and deleting the 8 potential polyadenylation sites and 12 mRNA destabilizing sequence motifs that were present. The codons were also optimized to enhance translation in dicots. The reconstructed vip3Ba gene was cloned between the T-DNA borders of the binary vector, pART27 (Fig. 2), and electroporated into A. tumefaciens AGL1.
Schematic diagram of the Agrobacterium binary vector T-DNA; LB, RB left and right borders of Agrobacterium T-DNA, respectively. The antibiotic selection gene neomycin phosphotransferase II (nptII) from E. coli was flanked by the S1 promoter derived from segment 1 of the subterranean clover stunt virus (SCSV) genome and segment 3 (S3) 3′ end while the optimized coding region of the vip3Ba gene was flanked by the Arabidopsis thaliana small subunit (AraSSU) promoter and Nicotiana tabacum small subunit (TobSSU) 3′ end
The reconstructed vip3Ba gene was transformed into cowpea following a modification of the protocol described by Popelka et al. (2006). A band of the expected size for Vip3Ba (~75 kDa) was detected by western blot in extracts from seven lines namely V9, V24, V25, V43, V56, V87 and V107. An additional, non-specific band of ~22 kDa was also observed in protein extracts from all cowpea lines including the non-transformed negative control. A representative blot of total soluble protein (TSP) extracts from T1 progeny derived from lines V9, V24, V25, V43, V56, V87 and V107 showing average expression in these lines is displayed in Fig. 3. The concentration of Vip3Ba protein in the extracts was estimated by visual comparison with known levels (1, 3, 10, 30 and 100 ng) of E. coli-expressed Vip3Ba protein. The limit of detection of Vip3Ba protein was found to be 1.5 ng, which is equivalent to 37.5 ng/mg TSP (data not shown). Based on this, the levels of Vip3Ba protein expression in the seven lines ranged between 0.25 and 5.0 µg/mg TSP.
Western blots using a monoclonal antibody to Vip3Ba showing the levels of Vip3Ba in seven T1 cowpea lines using varying levels of Vip3Ba expressed in E. coli as standards. In both blots, Lane M is the protein molecular weight ladder. Lanes L1–L5 represent E. coli expressed Vip3Ba loaded at 100, 50, 25, 12.5 and 6.25 ng per lane, respectively. Blot A includes protein (40 µg/lane) from cowpea lines 9, 24, 25 and 43 while Blot B includes protein (40 µg/lane) from lines 56, 87 and 107
Transgenic cowpea leaves expressing Vip3Ba protein were toxic to Maruca larvae
A replicated series of bioassays was carried out to test the efficacy of transgenic lines expressing the Vip3Ba toxin on the growth and development of MPB larvae. 16 replicate leaf samples from four Vip3Ba-expressing cowpea lines (with levels ranging between 155 and 895 ng/mg TSP) were fed to MPB larvae. The bioassay was run over a 10-day period and was followed by measuring MPB mortality and the weight of any surviving larvae. As controls, samples were also taken from leaves of the positive control Cry1Ab-line (709A) (which has 260 ng Cry1Ab protein per mg TSP) and from the non-transgenic negative control line (IT86D). The average weight of larvae fed on leaf samples from IT86D ranged between 23 and 26 mg after 10 days’ growth and no mortality was observed. Most of the larvae fed on IT86D leaves had progressed to the fifth and final instar stage by the end of the trial. When larvae were fed on leaves from the positive control line, 709A, expressing Cry1Ab, the mortality was 100% as expected. There was also 100% mortality of larvae feeding on leaves of all four transgenic cowpea lines expressing Vip3Ba protein (Table 2; Fig. 4). Leaf damage on the Vip3Ba-transgenic lines, as well as the positive control, was negligible and most of the leaf discs remained fully intact (Fig. 4). In contrast, leaf damage on line IT86D was severe with abundant frass observed on the leaf disks indicating that the insects feeding on these plants were developing normally (Fig. 4).
Leaf damage after 10 days of feeding by Maruca larvae on non-transgenic line IT86D and transgenic lines expressing Vip3Ba or Cry 1Ab protein. a Line 709A: (positive control expressing Cry 1Ab protein), b line V24 (425 ng Vip3Ba/mg TSP), c line V25 (155 ng Vip3Ba/mg TSP), d line V43 (895 ng Vip3Ba/mg TSP), e line V87 (365 ng Vip3Ba/mg TSP) and f line IT86D (0 ng Vip3Ba/mg TSP)
Discussion
Cowpea production worldwide is constrained by several insect pests, however, the Maruca pod borer (MPB) is considered one of the most devastating. The absence of genes for resistance to Maruca in cowpea germplasm has precluded the use of conventional plant breeding as a means to control this pest (Fatokun 2002). As such, genetic engineering appears to be the most viable control option for developing host plant resistance. The aim of this work was to identify a vip3 gene in a collection of Australian Bt strains that encoded a toxin active against the MPB, generate transgenic cowpeas expressing Vip3 toxin and determine whether they were resistant to MPB. The vip3 genes were targeted as they are known to have insecticidal activity against certain lepidopteran pests (Estruch et al. 1996; Beard et al. 2008). Furthermore, transgenic cotton expressing a vip3A gene was protected against the cotton boll weevil (CBW) (Lepidoptera) (Wu et al. 2011). Elsewhere, transgenic cotton (VipCot) expressing both Vip3A and Cry1Ab was shown to be more effective in controlling two lepidopteran pests compared to cotton expressing a single Cry gene (Cry1Ac) (Bommireddy et al. 2011). In maize, a pyramided (Vip3A and Cry1Ab) hybrid was shown to control key lepidopteran pests (Burkness et al. 2010). Similarly, the pyramiding of a drought tolerance gene with a Cry2Aa2 gene led to insect resistance and enhanced water efficiency in pepper (Zhu et al. 2015).
To increase the probability of identifying a vip3 gene product with activity against MPB, a diverse range of Vip3-encoded proteins were targeted. The selection of the target protein 2s was based on sequence analysis of Vip3 amino acid sequences in the Bt database. Approximately 60% of sequences in the Bt database encoded Vip3Aa proteins with 95% amino acid identity and Vip3Aa35 was arbitrarily chosen as one target. Using Vip3Aa35 as a reference, four additional Vip3 target proteins, namely Vip3Af1, Vip3Ag, Vip3Ca2 and Vip3Ba1, were selected whose amino acid sequences showed between 90 and 60% identity to vip3Aa35.
The screening of the Australian Bt collection for genes encoding the target Vip3 proteins revealed that it was dominated by vip3A genes (35%), with a small number of vip3B genes (5%) and no vip3C genes. In other studies, the incidence of vip3A genes ranged from 18 to 87% (Doss et al. 2002; Bhalla et al. 2005; Liu et al. 2007; Beard et al. 2008; Hernandez-Rodriguez et al. 2009; Yu et al. 2011b). The abundance of vip3 genes observed in the current study was also generally in accordance with the Bt database (Crickmore et al. 2014) where the majority of vip3 genes belong to the A family, with fewer members in the B family and very few in family C.
In this Australian Bt collection vip3Aa35, vip3Af1, vip3Ag and vip3Ba1 were identified whereas no Bt isolate containing vip3Ca2 was found. The full coding sequences of three of the five target vip genes (vip3Aa35, vip3Ag and vip3Ba1) were amplified and cloned into a protein expression vector. It was necessary to chemically synthesise the coding sequences of vip3Af1 and vip3Ca2 because they were either unable to be cloned (vip3Af1) or were not found in the Bt collection (vip3Ca2). Following cloning, the five vip3 genes were expressed in E. coli. Based on the amino acid sequence of the encoded proteins, the expected sizes of the proteins were around 90 kDa. This was consistent with the sizes of Vip3 proteins reported by others (Estruch et al. 1996; Beard et al. 2008; Yu et al. 2011b; Palma et al. 2012; Hernandez et al. 2013). Although the sizes of the five expressed Vips were predicted to be approximately 90 kDa, the major band observed in the protein extracts in this study was approximately 75 kDa. This was not entirely unexpected as it is known that the migration of proteins electrophoresed through SDS–PAGE is influenced by many factors and does not always provide an accurate reflection of their true molecular weight (Jong et al. 1978). For example, one possible reason why the putative Vip proteins resolved at 75 kDa is due to a high pH in the Tris–glycine buffer system accelerates the rate of protein migration on SDS–PAGE (Schagger and von Jagow 1987; Liu and Chang 2010). However, to provide more definitive evidence that the 75 kDa bands were indeed Vips, the protein bands were excised and analysed by MS/MS. The analysis revealed a strong probability that the bands were the expected Vips.
The Vip3Aa35, Vip3Af1, Vip3Ag and Vip3Ca2 were mainly present in the insoluble protein fraction but Vip3Ba1 was found in both soluble and insoluble fractions. A protein solubility prediction (Wilkinson and Harrison 1991) carried out for the Vip3 protein sequences revealed that the chances of full solubility for most of these proteins when over-expressed in E. coli were low. The solubility scores for Vip3Aa35, Vip3Af, Vip3Ag and Vip3Ca2 ranged from 37 to 48%. In contrast, Vip3Ba was predicted to have a 59.1% chance of being soluble. Further, when Rang et al. (2005) and Beard et al. (2008) over-expressed Vip3Ba1 and Vip3Bb2 proteins in E. coli, they were shown to be present in the soluble and insoluble fractions, respectively. In other studies, Palma et al. (2013) reported that Vip3Aa45 and Vip3Ag4 were present in both the soluble and insoluble fractions, while Hernandez-Martinez et al. (2013) and Escudero et al. (2014) extracted soluble forms of Vip3Aa, Vip3Ab, Vip3Ad, Vip3Ae and Vip3Af.
High molecular weight bands and some lower MW bands were also found in some of the Vip3 protein extracts. The high molecular bands are possibly aggregates, a phenomenon commonly reported during the overexpression of recombinant protein in E. coli (Lebendiker and Danieli 2014). This can sometimes be overcome by the use of fusion proteins and buffers or solvents to obtain stable proteins and proper protein folding (Bondos and Bicknell 2003; Sorensen and Mortensen 2005). It is possible that the low molecular weight bands reflect cleavage products of the recombinant protein. Andberg et al. (2007) reported that proteins with purification tags were easily cleaved in the presence of a buffer and metal salts. This is likely with the Vip proteins in this work as they contained His tags and were combined with a buffer that contained Tris–HCl and sodium salts.
The Maruca bioassays showed that Vip proteins from the 3A and 3C families (Vip3Aa35, Vip3Af1, Vip3Ag, Vip3Ca2) had no effect on caterpillar growth, and the larvae developed into fifth instars. Palma et al. (2012) also reported that Vip3C toxin had little or no effect on seven other lepidopteran pests. However, several studies have reported growth-inhibiting effects of five different Vip3A proteins on selected lepidopteran pests (Estruch et al. 1996; Chakroun et al. 2012; Hernandez-Martinez et al. 2013). In this study, Vip3Ba toxin inhibited the growth of MPB larvae by 90% when exposed to 3 µg/g diet. These results are consistent with, and extend the data of Rang et al. (2005), who demonstrated that Vip3Ba impaired the larval growth of two other lepidopteran species (Ostrinia nubilalis and Plutella xylostella).
In general, insecticidal proteins need to be expressed at relatively high levels in their plant hosts in order to effectively control the insect target (Gatehouse 2008). One limitation of using native Bt genes is that they are expressed poorly in higher eukaryotes (Schuler et al. 1998; Perlak et al. 2001). This is most likely due to the fact these bacterial genes are AT-rich and contain cryptic polyadenylation signals and mRNA destabilizing sequence motifs; characteristics that can negatively affect mRNA processing in plants (Estruch et al. 1997; Jouanin et al. 1998). In order to maximise vip3Ba gene expression in transgenic cowpea, the coding sequence of the bacterially-derived vip3Ba gene was modified using the strategy described by Perlak et al. (1991). This involved increasing the GC content of the coding sequence and deleting the polyadenylation and mRNA destabilizing sequences without altering the amino acid sequence. Also, codon usage was optimized to enhance translation in dicotyledonous plants by using plant-preferred codons. Using a similar strategy, Das et al. (2016) constructed a plant codon-optimized chimeric Cry1Aabc gene for expression in pigeon pea. Likewise, high level expressions of optimized cry1Ac, cry1Ab and cry1C genes have been reported for engineering resistance to lepidopteran insect pests in transgenic cotton, tomato and tobacco, respectively (Perlak et al. 1990, 1991; van der Salm et al. 1994).
When cowpeas were transformed with a reconstructed vip3Ba gene, four lines expressing different levels of Vip3Ba (ranging from 155 to 895 ng/mg TSP) were tested for their efficacy against MPB larvae. Despite the varied levels of Vip3Ba between plants, there was 100% mortality of M. vitrata larvae in all transgenic lines tested. This shows that Vip3Ba levels of 155 ng/mg TSP are sufficient to cause Maruca larvae death, and that this toxin is highly effective at relatively low doses. In other research, expression of Bt endotoxin (Cry1A) as low as 25 ng/mg TSP was sufficient to cause 100% mortality in Helicoverpa armigera (Das et al. 2016).
The results presented here demonstrate for the first time that the growth of MPB larvae is severely inhibited by all levels of Vip3Ba protein tested, both in vitro and in planta. Further, we have shown that transgenic cowpea lines expressing Vip3Ba protein were completely protected against MPB larvae. Thus, it is proposed that the vip3Ba gene is an attractive candidate to complement cry genes in the development of Bt cowpeas resistant to MPB, thereby contributing to an effective strategy for insect resistance management. This combination would considerably reduce the likelihood of development of resistance in MPB, help to increase yield and income, and reduce the dependency on pesticides.
References
Acharjee S, Sarma BK, Kumar PA, Olsen K, Mahon R, Moar WJ, Moore A, Higgins TJV (2010) Transgenic chickpeas (Cicer arietinum L.) expressing a sequence-modified cry2Aa gene. Plant Sci 178:333–339
Andberg M, Jantti J, Heilimo S, Pihkala P, Paananen A, Koskinen AMP, Soderlund H, Linder MB (2007) Cleavage of recombinant proteins at poly-His sequences by Co(II) and Cu(II). Protein Sci 16:1751–1761
Beard CE, Court L, Boets A, Mourant R, Van Rie J, Akhurst RJ (2008) Unusually high frequency of genes encoding vegetative insecticidal proteins in an Australian Bacillus thuringiensis collection. Curr Microbiol 57:195–199
Bhalla R, Dalal M, Panguluri SK, Jagadish B, Mandaokar AD, Singh AK, Kumar PA (2005) Isolation, characterization and expression of a novel vegetative insecticidal protein gene of Bacillus thuringiensis. FEMS Microbiol Lett 243:467–472
Bommireddy PL, Leonard BR, Temple J, Price P, Emfinger K, Cook D, Hardke JT (2011) Field performance and seasonal efficacy profiles of transgenic cotton lines expressing Vip3A and VipCot against Helicoverpa zea (Boddie) and Heliothis virescens (F.). J Cotton Sci 15:251–259
Bondos SE, Bicknell A (2003) Detection and prevention of protein aggregation before, during, and after purification. Anal Biochem 316:223–231
Boukar O, Bhattacharjee R, Fatokun C, Kumar PL, Gueye B (2013) Cowpea. In: Mohar S, Upadhyaya HD, Bisht IS (eds) Genetic and genomic resources of grain legume improvement. Elsevier, Amsterdam, pp 137–156
Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal Biochem 72:248–254
Bravo A, Soberon M, Gill SS (2005) Bacillus thuringiensis: mechanisms and use. Comp Mol Insect Sci 6:175–205
Burkness EC, Dively G, Patton T, Morey AC, Hutchison WD (2010) Novel Vip3A Bacillus thuringiensis (Bt) maize approaches high-dose efficacy against Helicoverpa zea (Lepidoptera: Noctuidae) under field conditions. Implications for resistance management. GM Crops 1(5):337–343
Chakraborty J, Sen S, Ghosh P, Sengupta A, Basu D, Das S (2016) Homologous promoter derived constitutive and chloroplast targeted expression of synthetic cry1Ac in transgenic chickpea confers resistance against Helicoverpa armigera. Plant Cell Tiss Organ Cult 125:521–535
Chakroun M, Bel Y, Caccia S, Abdelkefi-Mesrati L, Escriche B, Ferre J (2012) Susceptibility of Spodoptera frugiperda and S. exigua to Bacillus thuringiensis Vip3Aa insecticidal protein. J Invertebr Pathol 110:334–339
Cottrell JS (2011) Protein identification using MS/MS data. J Proteomics 74:1842–1851
Crickmore N, Baum J, Bravo A, Lereclus, D, Narva K, Sampson K, Schnepf E, Sun M, Zeigler DR (2014) Bacillus thuringiensis toxin nomenclature. http://www.btnomenclature.info/. Accessed 22 Sept 2015
Das A, Datta S, Sujayanand GK, Kumar M, Singh AK, Arpan, Shukla A, Ansari J, Kumar M, Faruqui L, Thakur S, Kumar PA, Singh NP (2016) Expression of chimeric Bt gene, Cry1Aabc in transgenic pigeonpea (cv. Asha) confers resistance to gram pod borer (Helicoverpa armigera Hubner.). Plant Cell Tiss Organ Cult 127:705–715
Doss VA, Kumar KA, Jayakumar R, Sekar V (2002) Cloning and expression of the vegetative insecticidal protein (vVp3V) gene of Bacillus thuringiensis in Escherichia coli. Protein Expression Purif 26:82–88
Escudero IR, Banyuls N, Bel Y, Maeztu M, Escriche B, Munoz D, Caballero P, Ferre J (2014) A screening of five Bacillus thuringiensis Vip3A proteins for their activity against lepidopteran pests. J Invertebr Pathol 117:51–55
Estruch JJ, Warren GW, Mullins MA, Nye GJ, Craig JA, Koziel MG (1996) Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc Natl Acad Sci USA 93:5389–5394
Estruch JJ, Carozzi NB, Desai N, Duck NB, Warren GW, Koziel MG (1997) Transgenic plants: an emerging approach to pest control. Nat Biotechnol 15:137–141
FAOSTAT (2016). http://faostat.fao.org/beta/en/#data/QC
Fatokun CA (2002) Breeding cowpea for resistance to insect pests: attempted crosses between cowpea and Vigna vexillate. In: Fatokun CA, Tarawali SA, Singh BB, Kormawa PM, Tamo M (eds) Challenges and opportunities for enhancing sustainable cowpea production. Proceedings of the World Cowpea Conference III. IITA, Ibadan, pp 52–61
Gatehouse JA (2008) Biotechnological prospects for engineering insect-resistant plants. Plant Physiol 146:881–887
Gibson DG (2011) Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361
Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20:1203–1207
Gouffon C, Van Vliet A, Van Rie J, Jansens S, Jurat-Fuentes JL (2011) Binding sites for Bacillus thuringiensis Cry2Ae toxin on heliothine brush border membrane vesicles are not shared with Cry1A, Cry1F, or Vip3A toxin. Appl Environ Microbiol 77:3182–3188
Hall AE, Cisse N, Thiaw S, Elawad HOA, Ehlers JD, Ismail AM, Fery RL, Roberts PA, Kitch LW, Murdock LL, Boukar O, Phillips RD, McWatters KH (2003) Development of cowpea cultivars and germplasm by the Bean/Cowpea CRSP. Field Crops Res 82:103–134
Harwood RR (1979) Small farm development: understanding and improving farming systems in the humid tropics. International Agricultural Development Service, Westview Press, Inc., Boulder
Hernandez D, Rodriguez-Cabrera L, Valdes R, Moran I, Tellez P, Riveron A, Ramos Y, Gomez L, Ayra-Pardo C (2013) Bacillus thuringiensis Vip3Aa1 expression and purification from E. coli to be determined in seeds and leaves of genetically-modified corn plants. J Agron 12:153–167
Hernandez-Martinez P, Hernandez-Rodriquez CS, Van Rie J, Escriche B, Ferre J (2013) Insecticidal activity of Vip3Aa, Vip3Ad, Vip3Ae, and Vip3Af from Bacillus thuringiensis against lepidopteran corn pests. J Invertebr Pathol 113:78–81
Hernandez-Rodriguez CS, Boets A, Van Rie J, Ferre J (2009) Screening and identification of vip genes in Bacillus thuringiensis strains. J Appl Microbiol 107:219–225
Higgins TJV, Gollasch S, Molvig L et al (2012) Insect-protected cowpeas using gene technology. In: Boukar O, Coulibaly O, Fatokun CA et al (eds) Innovative research along the cowpea value chain. Proceedings of the Fifth World Cowpea Conference on improving livelihoods in the cowpea value chain through advancement in science. Saly, Senegal 27 September–1 October 2010. International Institute of Tropical Agriculture. Ibadan, pp 131–137
Jackai LEN (1995) The legume pod borer Maruca testulalis, and its principal host plant, Vigna unguiculata (L.) Walp.—use of selective insecticide sprays as an aid in the identification of useful levels of resistance. Crop Prot 14:299–306
Jackai LEN, Daoust RA (1986) Insect pests of cowpeas. Annu Rev Entomol 31:95–119
Jackai LEN, Raulston JR (1988) Rearing the legume pod borer, Maruca testulalis, Geyer (Lepidoptera: Pyralidae) on artificial diet. Trop Pest Manag 34:168–172
Jacob C, Carrasco B, Schwember AR (2016) Advances in breeding and biotechnology of legume crops. Plant Cell Tiss Organ Cult 127:561–584
Jong de WW, Zweers A, Cohen LH (1978) Influence of single amino acid substitutions on electrophoretic mobility of sodium dodecyl sulfate-protein complexes. Biochem Biophys Res Commun 82:532–539
Jouanin L, Bonade-Bottino M, Girard C, Morrot G, Giband M (1998) Transgenic plants for insect resistance. Plant Sci 131:1–11
Kaur A, Sharma M, Sharma C, Kaur H, Kaur N, Sharma S, Arora R, Singh I, Sandhu JS (2016) Pod borer resistant transgenic pigeon pea (Cajanus cajan L.) expressing cry1Ac transgene generated through simplified Agrobacterium transformation of pricked embryo axes. Plant Cell Tiss Organ Cult 127:717–727
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics. 23, 2947–2948. http://www.ebi.ac.uk/Tools/services/web/toolform.ebi?tool=clustalw2
Lebendiker M, Danieli T (2014) Production of prone-to-aggregate proteins. FEBS Lett 588:236–246
Lee MK, Walters FS, Hart H, Palekar N, Chen J-S (2003) The mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3A differs from that of Cry1Ab δ-endotoxin. Appl Environ Microbiol 69:4648–4657
Liu B-Y, Chang J-Y (2010) Rapid and irreversible reduction of protein disulfide bonds. Anal Biochem 405:67–72
Liu J, Song F, Zhang J, Liu R, He K, Tan J, Huang D (2007) Identification of vip3A-type genes from Bacillus thuringiensis strains and characterization of a novel vip3A-type gene. Lett Appl Microbiol 45:432–438
Machuka J, Adesoye A, Obembe OO (2000) Regeneration and genetic transformation in cowpea. In: Fatokun CA, Tarawali SA, Singh BB, Kormawa PM, Tamo M (eds) Challenges and opportunities for enhancing sustainable cowpea production. Proceedings of the World Cowpea Conference III. IITA, Ibadan, pp 185–196
Margam VM, Coates BS, Ba MN, Sun W, Binso-Dabire CL, Baoua I, Ishiyaku MF, Shukle JT, Hellmich RL, Covas FG, Ramasamy S, Armstrong J, Pittendrigh BR, Murdock LL (2011) Geographic distribution of phylogenetically-distinct legume pod borer, Maruca vitrata (Lepidoptera: Pyraloidea: Crambidae). Mol Biol Rep 38:893–903
Mohammed BS, Ishiyaku MF, Abdullahi US, Katung MD (2014) Response of transgenic Bt cowpea lines and their hybrids under field conditions. J Plant Breed Crop Sci 6:91–96
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Murdock LL, Coulibaly O, Higgins TJV, Huesing JE, Ishiyaku M, Sithole-Niang I (2008) Cowpea. In: Kole C, Hall TC (eds) Compendium of transgenic crop plants: transgenic legume grains and forages. Blackwell Publishing, Oxford, pp 23–56
Nkongolo KK, Bokosi J, Malusi M, Vokhiwa Z, Mphepo M (2009) Agronomic, culinary, and general characterization of selected cowpea elite lines using farmers’ and breeder’s knowledge: a case study from Malawi. Afr J Plant Sci 3(7):147–156
Palma L, Hernandez-Rodriguez CS, Maeztu M, Hernandez-Martinez P, de Escudero IR, Escriche B, Munoz D, Van Rie J, Ferre J, Caballero P (2012) Vip3C, a novel class of vegetative insecticidal proteins from Bacillus thuringiensis. Appl Environ Microbiol 78:7163–7165
Palma L, de Escudero IR, Maeztu M, Caballero P, Munoz D (2013) Screening of vip genes from a Spanish Bacillus thuringiensis collection and characterization of two Vip3 proteins highly specific to five lepidopteran crop pests. Biol Control 66:141–149
Perlak FJ, Deaton RW, Armstrong TA, Fuchs RL, Sims SR, Greenplate JT, Fischhoff DA (1990) Insect resistant cotton plants. Nat Biotechnol 8:939–943
Perlak FJ, Fuchs RL, Dean DA, McPherson SL, Fischhoff DA (1991) Modification of the coding sequence enhances plant expression of insect control protein genes. Proc Natl Acad Sci USA 88:3324–3328
Perlak FJ, Oppenhuizen M, Gustafson K, Voth R, Sivasupramaniam S, Heering D, Carey B, Ihrig RA, Roberts JK (2001) Development and commercial use of Bollgard cotton in the USA—early promises versus today’s reality. Plant J 27:489–501
Pimentel D, Acquay H, Biltonen M, Rice P, Silva M, Nelson J, Lipner V, Giordano S, Horowitz A, D’Amore M (1992) Environmental and economic costs of pesticide use. Bioscience 42:750–760
Popelka JC, Gollasch S, Moore A, Molvig L, Higgins TJV (2006) Genetic transformation of cowpea (Vigna unguiculata L.) and stable transmission of the transgenes to progeny. Plant Cell Rep 25:304–312
Qaim M (2009) The economics of genetically modified crops. Annu Rev Resour Econ 1:665–693
Rang C, Gil P, Neisner N, Rie Van J, Frutos R (2005) Novel Vip3-related protein from Bacillus thuringiensis. Appl Environ Microbiol 71:6276–6281
Roush R (1998) Two-toxin strategies for management of insecticidal transgenic crops: can pyramiding succeed where pesticide mixtures have not? Phil Trans R Soc B 353:1777–1786
Schagger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379
Schuler TH, Poppy GM, Kerry BR, Denholm I (1998) Insect-resistant transgenic plants. Trends Biotechnol 16:168–175
Schunmann PHD, Llewellyn DJ, Surin B, Boevink P, De Feyter RC, Waterhouse PM (2003) A suite of novel promoters and terminators for plant biotechnology. Funct Plant Biol 30:443–452
Singh SR, Van Emden HF (1979) Insect pests of grain legumes. Annu Rev Entomol 24:255–278
Sorensen HP, Mortensen KK (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact 4:1
Srinivasan R (2008) Susceptibility of legume pod borer (LPB), Maruca vitrata to δ-endotoxins of Bacillus thuringiensis (Bt) in Taiwan. J Invertebr Pathol 97:79–81
Studier FW (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41:207–234
Tabe LM, Wardley-Richardson T, Ceriotti A, Aryan A, McNabb W, Moore A, Higgins TJV (1995) A biotechnological approach to improving the nutritive value of alfalfa. J Anim Sci 73:2752–2759
Tamo M, Ekesi S, Maniania NK, Cherry A (2003) Biological control, a non-obvious component of IPM for cowpea. In: Neuenschwander P, Borgemeister C, Langewald J (eds) Biological control in IPM systems in Africa. CABI Publishing, Wallingford, pp 295–309
Taylor TA (1968) The pathogenicity of Bacillus thuringiensis var. thuringiensis Berliner for larvae of Maruca testulalis Geyer. J Invertebr Pathol 11:386–389
Timko MP, Singh BB (2008) Cowpea, a multifunctional legume. In: Genomics of tropical crop plants. Springer, New York, pp 227–258
Van der Salm T, Bosch D, Honee G, Feng L, Munsterman E, Bakker P, Stiekema WJ, Visser B (1994) Insect resistance of transgenic plants that express modified Bacillus thuringiensis cry1A(b) and cry1C genes: a resistance management strategy. Plant Mol Biol 26:51–59
Warren GW (1997) Vegetative insecticidal proteins: novel proteins for control of corn pests. In: Carozzi N, Koziol M (eds) Advances in insect control: the role of transgenic plants. Taylor and Francis, London, pp 109–121
Whitehouse MEA, Wilson LJ, Constable GA (2007) Target and non-target effects on the invertebrate community of Vip cotton, a new insecticidal transgenic. Aust J Agric Res 58:273–285
Wilkinson DL, Harrison RG (1991) Predicting the solubility of recombinant proteins in Escherichia coli. Nat Biotechnol 9:443–448
Wilson FD, Flint HM, Deaton WR, Fischhoff DA, Perlak FJ, Armstrong TA, Fuchs RL, Berberich SA, Parks NJ (1992) Resistance of cotton lines containing a Bacillus thuringiensis toxin to pink bollworm (Lepidoptera: Gelechiidae) and other insects. J Econ Entomol 84:1516–1521
Wu J, Luo X, Zhang Z, Shi Y, Tian Y (2011) Development of insect-resistant transgenic cotton with chimeric TVip3A* accumulating in chloroplasts. Transgenic Res 20:963–973
Yu X, Liu T, Liang X, Tang C, Zhu J, Wang S, Li S, Deng Q, Wang L, Zheng A, Li P (2011a) Rapid detection of vip1-type genes from Bacillus cereus and characterization of a novel vip binary toxin gene. FEMS Microbiol Lett 325:30–36
Yu X, Zheng A, Zhu J, Wang S, Wang L, Deng Q, Li S, Liu H, Li P (2011b) Characterization of vegetative insecticidal protein vip genes of Bacillus thuringiensis from Sichuan Basin in China. Curr Microbiol 62:752–757
Zhu Z, Xu X, Cao B, Chen C, Chen Q, Xiang C, Chen G, Lei J (2015) Pyramiding of AtEDT1/HDG11 and Cry2Aa2 into pepper (Capsicum annuum L.) enhances drought tolerance and insect resistance without yield decrease. Plant Cell Tiss Organ Cult 120:919–932
Acknowledgements
This research was supported by Commonwealth Scientific and Industrial Research Organization (CSIRO) and the Australian Government (AusAID) through a scholarship at Queensland University of Technology (QUT). The authors thank Dan Pikler for cloning of the cry2Aa gene, Dan Llewellyn for providing a positive control for the vip3Aa gene and Lisa Molvig for her technical support and advice.
Author information
Authors and Affiliations
Contributions
TJVH and RH conceived and designed the study. BB, SG and AM performed the plant based experiments. BB, SG, WJ and JA performed the insect based experiments. BB, RH and TJVH analysed and interpreted the results. BB, RH, TW and TJVH prepared the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Sergio J Ochatt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bett, B., Gollasch, S., Moore, A. et al. Transgenic cowpeas (Vigna unguiculata L. Walp) expressing Bacillus thuringiensis Vip3Ba protein are protected against the Maruca pod borer (Maruca vitrata). Plant Cell Tiss Organ Cult 131, 335–345 (2017). https://doi.org/10.1007/s11240-017-1287-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-017-1287-3