Abstract
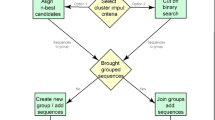
This work explains synthesis of protein structures based on the unsupervised learning method known as clustering. Protein structure prediction was performed for different crab and egg datasets with inputs collected from the Protein Data Bank (PDB ID: 3LIG, 2W3Z, 3ZVQ, 2KLR and 2YIZ). The three-dimensional protein structure was merged together with the filtering instances inbuilt in data mining techniques known as MergeSets. The problem description in this proposed methodology, referred to as attribute-related cluster sequence analysis, is to identify a good working algorithm for clustering of protein structures by comparing four existing algorithms: k-means, expectation maximization, farthest first and COBWEB. Experiments are conducted with the BioWeka data mining tool, Modeler 9.15 and the PyMOL tool with scripts using the Python programming language. This paper shows that the expectation maximization algorithm is the best for structured protein clustering, and this will also pave the way for identifying better algorithms for supervised learning methods.










Similar content being viewed by others
References
Vignesh U (2013) Implementing efficient DNA matching using suffix tree. Eng Sci Int Res J 1:170–172
Vignesh U, Sivakumar M (2013) Implementing high performance retrieval process by max-score ranking. IOSR J Comput Eng 8:28–33
Vignesh U, Senthilraja P (2013) MashQL Editor using Query Detection Algorithm. Eng Sci Int Res J 1:173–176
Vignesh U, Valarmathi P, Arun S (2013) Implementing clustering using CSI by K-means. Int J Eng Sci Innov Technol 2:568–573
Vignesh U, Parvathi R (2017) Clustering on structured proteins with filtering instances on Bioweka. J Eng Sci Technol 12:820–833
Vignesh U, Parvathi R (2017) Next generation sequencing data analysis software and methods: a survey. Int J Control Theory Appl 9:1–28
Vignesh S, Robert P, Vignesh U, Bharathidasan D, Rajasekaran S (2013) Implementing CURE to address scalability issue in social media. Int J Comput Eng Res 3:1–7
Birlutiu A, d’Alche-Buc F, Heskes T (2015) A Bayesian framework for combining protein and network topology information for predicting protein–protein interactions. IEEE Trans Comput Biol Bioinform 12(1):538–550
Song D, Chen J, Chen G, Li N, Li J, Fan J, Bu D, Li SC (2015) Parameterized BLOSUM matrices for protein alignment. IEEE Trans Comput Biol Bioinform 12(3):686–694
Tseng VA, Kao C-P (2005) Efficiently mining gene expression data via a novel parameterless clustering method. IEEE Trans Comput Biol Bioinform 2(1):355–365
Yang J, Wang W (2003) CLUSEQ: efficient and effective sequence clustering. In: 19th International Conference on Data Engineering, IEEE Computer Society Press, Los Alamitos, pp 101–112
Ng YK, Yin L, Ono H, Li SC (2015) Finding all longest common segments in protein structures efficiently. IEEE Trans Comput Biol Bioinform 12(3):644–655
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vignesh, U., Parvathi, R. 3D visualization and cluster analysis of unstructured protein sequences using ARCSA with a file conversion approach. J Supercomput 76, 4287–4301 (2020). https://doi.org/10.1007/s11227-018-2319-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-018-2319-4