Abstract
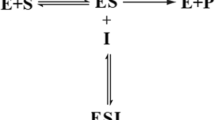
In the present paper, the mathematical modeling of highly sensitive enzyme biosensor kinetics is discussed. The standard method of inverting a Laplace transform according to the Heaviside expansion theorem is applied to solve the coupled nonlinear time-dependent reaction–diffusion equations for the Michaelis–Menten expression that describes the concentrations of the substrate and product within the enzymatic layer. The analytical expressions for the concentration of the substrate and product have been derived for all values of the rate constant. A numerical simulation is also reported using the MATLAB software program. Our analytical results are compared with our simulation results. The analytical results show good agreement with those obtained using numerical method.




Similar content being viewed by others
References
Gil ED, de Melo GR (2010) Electrochemical biosensors in pharmaceutical analysis. Braz J Pharm Sci 46:375–391
Djaalab E, Samar MEH, Zougar S, Kherrat R (2018) Electrochemical biosensor for the determination of amlodipine besylate based on gelatin-polyaniline iron oxide biocomposite film. Catalysts 8:233
Wanekaya AK, Chen W, Mulchandani A (2008) Recent biosensing developments in environmental security. J Environ Monit 10:703–710
Viswanathan S, Radecka H, Radecki J (2009) Electrochemical biosensors for food analysis. Monatsh Chem 140:891–899
Sethuraman V, Muthuraja P, Raj JA, Manisankar P (2016) A highly sensitive electrochemical biosensor for catechol using conducting polymer reduced graphene oxide–metal oxide enzyme modified electrode. Biosens Bioelectron 84:112–119
Zehani N, Fortgang P, Saddek LM, Baraket A, Arab M, Dzyadevych SV, Kherrat R, Jaffrezic-Renault N (2015) Highly sensitive electrochemical biosensor for bisphenol A detection based on a diazonium-functionalized boron-doped diamond electrode modified with a multi-walled carbon nanotubetyrosinase hybrid film. Biosens Bioelectron 74:830–835
Wang X, Hu J, Zhang G, Liu S (2014) Highly selective fluorogenic multianalyte biosensors constructed via enzyme-catalyzed coupling and aggregation-induced emission. J Am Chem Soc 136(28):9890–9893
Tang H, Yan F, Lin P, Xu J, Chan HLW (2011) Highly sensitive glucose biosensors based on organic electrochemical transistors using platinum gate electrodes modified with enzyme and nanomaterials. Adv Func Mater 21(12):2264–2272
Zheng Z, Zhou Y, Li X, Liu S, Tang Z (2011) Highly-sensitive organophosphorous pesticide biosensors based on nanostructured films of acetylcholinesterase and CdTe quantum dots. Biosens Bioelectron 26(6):3081–3085
Lu J, Drzal LT, Worden RM, Lee I (2007) Simple fabrication of a highly sensitive glucose biosensor using enzymes immobilized in exfoliated graphite nanoplatelets nafion membrane. Chem Mater 19(25):6240–6246
Lee D, Lee J, Kim J, Kim J, Na HB, Kim B, Kim HS (2005) Simple fabrication of a highly sensitive and fast glucose biosensor using enzymes immobilized in mesocellular carbon foam. Adv Mater 17(23):2828–2833
Lente G (2017) Advanced data analysis and modelling in chemical engineering. Reac Kinet Mech Cat 120(2):417–420
Yang KZ, Twaiq F (2017) Modelling of the dry reforming of methane in different reactors: a comparative study. Reac Kinet Mech Cat 122(2):853–868
Elizalde I, Trejo F, Muñoz JA, Torres P, Ancheyta J (2016) Dynamic modeling and simulation of a bench-scale reactor for the hydrocracking of heavy oil by using the continuous kinetic lumping approach. Reac Kinet Mech Cat 118(1):299–311
Izadbakhsh A, Khatami A (2014) Mathematical modeling of methanol to olefin conversion over SAPO-34 catalyst using the percolation concept. Reac Kinet Mech Cat 112(1):77–100
Marečić M, Jović F, Kosar V, Tomašić V (2011) Modelling of an annular photocatalytic reactor. Reac Kinet Mech Cat 103(1):19–29
Le TD, Lasseux D, Nguyen XP, Vignoles G, Mano N, Kuhn A (2017) Multi-scale modeling of diffusion and electrochemical reactions in porous micro-electrodes. Chem Eng Sci 173:153–167
Ingham J (2000) Chemical engineering dynamics: an introduction to modelling and computer simulation. Wiley-VCH, Weinheim
Yeo YK (2017) Chemical engineering computation with MATLAB, 1st edn. CRC Press, Boca Raton, FL
AL Malah K (2013) MATLAB numerical methods with chemical engineering applications. McGraw Hill, New York
Buzzi-Ferraris G, Manenti F (2013) Nonlinear systems and optimization for the chemical engineer: Solving numerical problems. Wiley-VCH, Weinheim
Ross G (1987) Computer programming examples for chemical engineers. Elsevier, Amsterdam
English BP, Min W, Van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS (2006) Ever-fluctuating single enzyme molecules: Michaelis–Menten equation revisited. Nat Chem Biol 2(2):87–94
Dóka É, Lente G (2012) Stochastic mapping of the Michaelis–Menten mechanism. J Chem Phys 136(5):054111
Kou SC, Cherayil BJ, Min W, English BP, Xie XS (2005) Single-molecule Michaelis–Menten equations. J Phys Chem B 109:19068–19081
Loney NW (2006) Applied mathematical methods for chemical engineers, 2nd edn. CRS Press, Boca Raton
Mathews JH, Howell RW (2012) Complex analysis for mathematics and engineering, 6th edn. Jones and Bartlett Learning, LLC, Bullington
MATLAB & Simulink, www.mathworks.com/help/matlab/ref/pdepe.html. Accessed 09 July, 2018
Villadsen J, Nielsen J, Liden G (2011) Bioreaction engineering principles. Springer, Dordrecht
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Djaalab, E., Samar, M.E.H. & Zougar, S. Mathematical modeling of the kinetics of a highly sensitive enzyme biosensor. Reac Kinet Mech Cat 126, 49–59 (2019). https://doi.org/10.1007/s11144-018-1516-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11144-018-1516-8