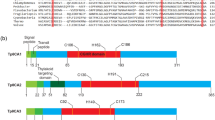
Abstract
Diatoms are unicellular algae and important primary producers. The process of carbon fixation in diatoms is very efficient even though the availability of dissolved CO2 in sea water is very low. The operation of a carbon concentrating mechanism (CCM) also makes the more abundant bicarbonate accessible for photosynthetic carbon fixation. Diatoms possess carbonic anhydrases as well as metabolic enzymes potentially involved in C4 pathways; however, the question as to whether a C4 pathway plays a general role in diatoms is not yet solved. While genome analyses indicate that the diatom Phaeodactylum tricornutum possesses all the enzymes required to operate a C4 pathway, silencing of the pyruvate orthophosphate dikinase (PPDK) in a genetically transformed cell line does not lead to reduced photosynthetic carbon fixation. In this study, we have determined the intracellular location of all enzymes potentially involved in C4-like carbon fixing pathways in P. tricornutum by expression of the respective proteins fused to green fluorescent protein (GFP), followed by fluorescence microscopy. Furthermore, we compared the results to known pathways and locations of enzymes in higher plants performing C3 or C4 photosynthesis. This approach revealed that the intracellular distribution of the investigated enzymes is quite different from the one observed in higher plants. In particular, the apparent lack of a plastidic decarboxylase in P. tricornutum indicates that this diatom does not perform a C4-like CCM.





Similar content being viewed by others
References
Allen AE, Moustafa A, Montsant A, Eckert A, Kroth PG, Bowler C (2012) Evolution and functional diversification of fructose bisphosphate aldolase genes in photosynthetic marine diatoms. Mol Biol Evol 29:367–379
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Alvarez CE, Saigo M, Margarit E, Andreo CS, Drincovich MF (2013) Kinetics and functional diversity among the five members of the NADP-malic enzyme family from Zea mays, a C4 species. Photosynth Res 115:65–80
Anning T, Nimer N, Merrett MJ, Brownlee C (1996) Costs and benefits of calcification in coccolithophorids. J Mar Syst 9:45–56
Appleby G, Colbeck J, Holdsworth ES (1980) Beta carboxylation enzymes in marine phytoplankton and isolation and purification of pyruvate carboxylase from Amphidinium carterae (Dinophyceae). J Phycol 16:290–295
Apt KE, Zaslavkaia L, Lippmeier JC, Lang M, Kilian O, Wetherbee R, Grossman AR, Kroth PG (2002) In vivo characterization of diatom multipartite plastid targeting signals. J Cell Sci 115:4061–4069
Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou SG, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kröger N, Lau WWY, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS (2004) The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 306:79–86
Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD (1998) The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can J Bot (Revue Canadienne De Botanique) 76:1052–1071
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Beopoulos A, Cescut J, Haddouche R, Uribelarrea JL, Molina-Jouve C, Nicaud JM (2009) Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res 48:375–387
Birmingham BC, Colman B (1979) Measurement of carbon dioxide compensation points of freshwater algae. Plant Physiol 64:892–895
Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N (2008) Multiple pathways for sorting mitochondrial precursor proteins. Embo Rep 9:42–49
Bowes G (2011) Single-cell C4 photosynthesis in aquatic plants. In: Raghavendra AS, Sage RF (eds) C4 photosynthesis and related CO2 concentrating mechanisms, vol 32, Springer, Dordrecht, pp 63–80
Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kröger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jézéquel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Oudot-Le Secq MP, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van De Peer Y, Grigoriev IV (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244
Braun FJ, Hegemann P (1999) Direct measurement of cytosolic calcium and pH in living Chlamydomonas reinhardtii cells. Eur J Cell Biol 78:199–208
Brown NJ, Palmer BG, Stanley S, Hajaji H, Janacek SH, Astley HM, Parsley K, Kajala K, Quick WP, Trenkamp S, Fernie AR, Maurino VG, Hibberd JM (2010) C4 acid decarboxylases required for C4 photosynthesis are active in the mid-vein of the C3 species Arabidopsis thaliana, and are important in sugar and amino acid metabolism. Plant J 61:122–133
Buchanan BB (1991) Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/ thioredoxin system Perspective on its discovery, present status, and future development. Arch Biochem Biophys 288:1–9
Chastain CJ, Xu WX, Parsley K, Sarath G, Hibberd JM, Chollet R (2008) The pyruvate, orthophosphate dikinase regulatory proteins of Arabidopsis possess a novel, unprecedented Ser/Thr protein kinase primary structure. Plant J 53:854–863
Chastain CJ, Failing CJ, Manandhar L, Zimmerman MA, Lakner MM, Nguyen THT (2011) Functional evolution of C4 pyruvate, orthophosphate dikinase. J Exp Bot 62:3083–3091
Chauton MS, Winge P, Brembu T, Vadstein O, Bones AM (2013) Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol 161:1034–1048
Clement R, Dimnet L, Maberly SC, Gontero B (2016) The nature of the CO2-concentrating mechanisms in a marine diatom, Thalassiosira pseudonana. New Phytol 209:1417–1427
Clement R, Jensen E, Prioretti L, Maberly SC, Gontero B (2017) Diversity of CO2 concentrating mechanisms and responses to CO2 concentration in marine and freshwater diatoms. J Exp Bot 68:3925–3935
Consortium TU (2014) Activities at the universal protein resource (UniProt). Nucleic Acids Res 42:D191-D198
Davis A, Abbriano R, Smith SR, Hildebrand M (2017) Clarification of photorespiratory processes and the role of malic enzyme in diatoms. Protist 168:134–153
Descolas-Gros C, Oriol L (1992) Variations in carboxylase activity in marine phytoplankton cultures. Beta-carboxylation in carbon flux studies. Mar Ecol Prog Ser 85:163–169
Detarsio E, Maurino VG, Alvarez CE, Müller GL, Andreo CS, Drincovich MF (2008) Maize cytosolic NADP-malic enzyme (ZmCytNADP-ME): a phylogenetically distant isoform specifically expressed in embryo and emerging roots. Plant Mol Biol 68:355–367
Dittrich P, Campbell WH, Black CC (1973) Phosphoenolpyruvate carboxykinase in plants exhibiting crassulacean acid metabolism. Plant Physiol 52:357–361
Edwards GE, Voznesenskaya EV (2011) C4 photosynthesis: Kranz forms and single-cell C4 in terrestrial plants. In: Raghavendra AS, Sage RF (eds) C4 photosynthesis and related CO2 concentrating mechanisms, vol 32, Springer, Dordrecht, pp 29–61
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Giordano M, Beardall J, Raven JA (2005) CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol 56:99–131
Gould SB, Sommer MS, Kroth PG, Gile GH, Keeling PJ, Maier UG (2006) Nucleus-to-nucleus gene transfer and protein retargeting into a remnant cytoplasm of cryptophytes and diatoms. Mol Biol Evol 23:2413–2422
Granum E, Raven JA, Leegood RC (2005) How do marine diatoms fix 10 billion tonnes of inorganic carbon per year? Can J Bot (Revue Canadienne De Botanique) 83:898–908
Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, Otillar R, Poliakov A, Ratnere I, Riley R, Smirnova T, Rokhsar D, Dubchak I (2012) The genome portal of the department of energy joint genome institute. Nucleic Acids Res 40:D26-D32
Gruber A, Kroth PG (2014) Deducing intracellular distributions of metabolic pathways from genomic data. Methods Mol Biol 1083:187–211
Gruber A, Kroth PG (2017) Intracellular metabolic pathway distribution in diatoms and tools for genome-enabled experimental diatom research. Philos Trans R Soc B 372:20160402
Gruber A, Vugrinec S, Hempel F, Gould SB, Maier UG, Kroth PG (2007) Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol Biol 64:519–530
Gruber A, Weber T, Río Bártulos C, Vugrinec S, Kroth PG (2009) Intracellular distribution of the reductive and oxidative pentose phosphate pathways in two diatoms. J Basic Microbiol 49:58–72
Gruber A, Rocap G, Kroth PG, Armbrust EV, Mock T (2015) Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J 81:519–528
Guillard RR, Ryther JH (1962) Studies of marine planktonic diatoms 1. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can J Microbiol 8:229–239
Haimovich-Dayan M, Garfinkel N, Ewe D, Marcus Y, Gruber A, Wagner H, Kroth PG, Kaplan A (2013) The role of C4 metabolism in the marine diatom Phaeodactylum tricornutum. New Phytol 197:177–185
Hao GF, Chen HQ, Wang L, Gu ZN, Song YD, Zhang H, Chen W, Chen YQ (2014) Role of malic enzyme during fatty acid synthesis in the oleaginous fungus Mortierella alpina. Appl Environ Microbiol 80:2672–2678
Harrison PJ, Waters RE, Taylor FJR (1980) A broad spectrum artificial seawater medium for coastal and open ocean phytoplankton. J Phycol 16:28–35
Häusler RE, Hirsch HJ, Kreuzaler F, Peterhänsel C (2002) Overexpression of C4-cycle enzymes in transgenic C3 plants: a biotechnological approach to improve C3-photosynthesis. J Exp Bot 53:591–607
Heber U (1974) Metabolite exchange between chloroplasts and cytoplasm. Annu Rev Plant Physiol Plant Mol Biol 25:393–421
Herve V, Derr J, Douady S, Quinet M, Moisan L, Lopez PJ (2012) Multiparametric analyses reveal the pH-dependence of silicon biomineralization in diatoms. Plos One 7:e46722
Holdsworth ES, Colbeck J (1976) The pattern of carbon fixation in the marine unicellular alga Phaeodactylum tricornutum. Mar Biol 38:189–199
Hopkinson BM, Meile C, Shen C (2013) Quantification of extracellular carbonic anhydrase activity in two marine diatoms and investigation of its role. Plant Physiol 162:1142–1152
Izui K, Matsumura H, Furumoto T, Kai Y (2004) Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu Rev Plant Biol 55:69–84
Jenkins CLD, Hatch MD (1985) Properties and reaction mechanism of C4 leaf pyruvate,Pi dikinase. Arch Biochem Biophys 239:53–62
Jenkins CLD, Burnell JN, Hatch MD (1987a) Form of inorganic carbon involved as a product and as an inhibitor of C4 acid decarboxylases operating in C4 photosynthesis. Plant Physiol 85:952–957
Jenkins CLD, Harris RLN, McFadden HG (1987b) 3,3-dichloro-2-dihydroxyphosphinoylmethyl-2-propenoate, a new, specific inhibitor of phosphoenolpyruvate carboxylase. Biochem Int 14:219–226
Kaplan A, Reinhold L (1999) CO2 concentrating mechanisms in photosynthetic microorganisms. Annu Rev Plant Physiol Plant Mol Biol 50:539–570
Kikutani S, Nakajima K, Nagasato C, Tsuji Y, Miyatake A, Matsuda Y (2016) Thylakoid luminal theta-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci USA 113:9828–9833
Kilian O, Kroth PG (2005) Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J 41:175–183
Kitao Y, Harada H, Matsuda Y (2008) Localization and targeting mechanisms of two chloroplastic β-carbonic anhydrases in the marine diatom Phaeodactylum tricornutum. Physiol Plant 133:68–77
Kroth PG, Chiovitti A, Gruber A, Martin-Jézéquel V, Mock T, Parker MS, Stanley MS, Kaplan A, Caron L, Weber T, Maheswari U, Armbrust EV, Bowler C (2008) A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. Plos ONE 3:e1426
Kustka AB, Milligan AJ, Zheng HY, New AM, Gates C, Bidle KD, Reinfelder JR (2014) Low CO2 results in a rearrangement of carbon metabolism to support C4 photosynthetic carbon assimilation in Thalassiosira pseudonana. New Phytol 204:507–520
Lai LB, Tausta SL, Nelson TM (2002) Differential regulation of transcripts encoding cytosolic NADP-malic enzyme in C3 and C4 Flaveria species. Plant Physiol 128:140–149
Leegood RC (2002) C4 photosynthesis: principles of CO2 concentration and prospects for its introduction into C3 plants. J Exp Bot 53:581–590
Li YY, Xu JJ, Ul Haq N, Zhang H, Zhu XG (2014) Was low CO2 a driving force of C4 evolution: Arabidopsis responses to long-term low CO2 stress. J Exp Bot 65:3657–3667
Liaud MF, Lichtle C, Apt K, Martin W, Cerff R (2000) Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway. Mol Biol Evol 17:213–223
Matsuda Y, Hara T, Colman B (2001) Regulation of the induction of bicarbonate uptake by dissolved CO2 in the marine diatom, Phaeodactylum tricornutum. Plant Cell Environment 24:611–620
Matsuda Y, Nakajima K, Tachibana M (2011) Recent progresses on the genetic basis of the regulation of CO2 acquisition systems in response to CO2 concentration. Photosynth Res 109:191–203
McFadden GI (2001) Primary and secondary endosymbiosis and the origin of plastids. J Phycol 37:951–959
Moog D, Stork S, Zauner S, Maier UG (2011) In silico and in vivo investigations of proteins of a minimized eukaryotic cytoplasm. Genome Biol Evol 3:375–382
Nakajima K, Tanaka A, Matsuda Y (2013) SLC4 family transporters in a marine diatom directly pump bicarbonate from seawater. Proc Natl Acad Sci USA 110:1767–1772
Nelson DM, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B (1995) Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cycles 9:359–372
Nikolau BJ, Ohlrogge JB, Wurtele ES (2003) Plant biotin-containing carboxylases. Arch Biochem Biophys 414:211–222
Offermann S, Okita TW, Edwards GE (2011) Resolving the compartmentation and function of C4 photosynthesis in the single-cell C4 species Bienertia sinuspersici. Plant Physiol 155:1612–1628
Parsley K, Hibberd JM (2006) The Arabidopsis PPDK gene is transcribed from two promoters to produce differentially expressed transcripts responsible for cytosolic and plastidic proteins. Plant Mol Biol 62:339–349
Pedersen AG, Nielsen H (1997) Neural network prediction of translation initiation sites in eukaryotes: perspectives for EST and genome analysis. Ismb-97, Fifth International Conference on Intelligent Systems for Molecular Biology, Proceedings 5:226–233
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Ratledge C, Wynn JP (2002) The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol 51:1–51
Ray TB, Black CC (1976) Inhibition of oxalacetate decarboxylation during C4 photosynthesis by 3-mercaptopicolinic acid. J Biol Chem 251:5824–5826
Reinfelder JR (2011) Carbon concentrating mechanisms in eukaryotic marine phytoplankton. In: Carlson CA, Giovannoni SJ (eds) Annual review of marine science, vol 3, Annual Reviews, Palo Alto, pp 291–315
Reinfelder JR, Kraepiel AML, Morel FMM (2000) Unicellular C4 photosynthesis in a marine diatom. Nature 407:996–999
Reiskind JB, Bowes G (1991) The role of phosphoenolpyruvate carboxykinase in a marine macroalga with C4-like photosynthetic characteristics. Proc Natl Acad Sci USA 88:2883–2887
Riebesell U, Wolfgladrow DA, Smetacek V (1993) Carbon dioxide limitation of marine phytoplankton growth rates. Nature 361:249–251
Río Bártulos C (2007) Eine mitochondriale Teil-Glykolyse bei Phaeodactylum tricornutum und anderen Heterokonten: Nachweis der Kompartimentierung mittels dem “green fluorescent protein” und molekulare Phylogenie. Dissertation, Technische Universität Carolo-Wilhelmina zu Braunschweig
Roberts K, Granum E, Leegood RC, Raven JA (2007a) C3 and C4 pathways of photosynthetic carbon assimilation in marine diatoms are under genetic, not environmental, control. Plant Physiol 145:230–235
Roberts K, Granum E, Leegood RC, Raven JA (2007b) Carbon acquisition by diatoms. Photosynth Res 93:79–88
Rottberger J, Gruber A, Boenigk J, Kroth PG (2013) Influence of nutrients and light on autotrophic, mixotrophic and heterotrophic freshwater chrysophytes. Aquat Microb Ecol 71:179–191
Sage RF, Christin PA, Edwards EJ (2011) The C4 plant lineages of planet Earth. J Exp Bot 62:3155–3169
Sage RF, Sage TL, Kocacinar F (2012) Photorespiration and the evolution of C4 photosynthesis. In: Merchant SS (ed) Annual review of plant biology, vol 63, Annual Reviews, Palo Alto, pp 19–47
Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Samukawa M, Shen C, Hopkinson BM, Matsuda Y (2014) Localization of putative carbonic anhydrases in the marine diatom, Thalassiosira pseudonana. Photosynth Res 121:235–249
Schaeffer ML, Harper LC, Gardiner JM, Andorf CM, Campbell DA, Cannon EKS, Sen TZ, Lawrence CJ (2011) MaizeGDB: curation and outreach go hand-in-hand. Database 2011:bar022
Scheibe R (1987) NADP+-malate dehydrogenase in C3-plants: regulation and role of a light-activated enzyme. Physiol Plant 71:393–400
Shao H, Gontero B, Maberly SC, Jiang HS, Cao Y, Li W, Huang WM (2017) Responses of Ottelia alismoides, an aquatic plant with three CCMs, to variable CO2 and light. J Exp Bot 68:3985–3995
Sheen J (1991) Molecular mechanisms underlying the differential expression of Maize pyruvate, orthophosphate dikinase genes. Plant Cell 3:225–245
Sheen JY, Bogorad L (1987) Differential expression of C4 pathway genes in mesophyll and bundle sheath cells of greening Maize leaves. J Biol Chem 262:11726–11730
Shen C, Dupont CL, Hopkinson BM (2017) The diversity of carbon dioxide-concentrating mechanisms in marine diatoms as inferred from their genetic content. J Exp Bot 68:3937–3948
Smith CM, Duff SMG, Chollet R (1994) Partial purification and characterization of Maize-leaf pyruvate, orthophosphate dikinase regulatory protein: a low-abundance, mesophyll-chloroplast stromal protein. Arch Biochem Biophys 308:200–206
Sommer MS, Gould SB, Lehmann P, Gruber A, Przyborski JM, Maier UG (2007) Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol Biol Evol 24:918–928
Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. In: Proceedings of the International Conference on Intelligent Systems for Molecular Biology 6:175–182
Stucka R, Dequin S, Salmon JM, Gancedo C (1991) DNA-sequences in chromosome II and chromosome VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol General Genet 229:307–315
Tachibana M, Allen AE, Kikutani S, Endo Y, Bowler C, Matsuda Y (2011) Localization of putative carbonic anhydrases in two marine diatoms, Phaeodactylum tricornutum and Thalassiosira pseudonana. Photosynth Res 109:205–221
Tanaka R, Kikutani S, Mahardika A, Matsuda Y (2014) Localization of enzymes relating to C4 organic acid metabolisms in the marine diatom, Thalassiosira pseudonana. Photosynth Res 121:251–263
Taylor L, Nunes-Nesi A, Parsley K, Leiss A, Leach G, Coates S, Wingler A, Fernie AR, Hibberd JM (2010) Cytosolic pyruvate,orthophosphate dikinase functions in nitrogen remobilization during leaf senescence and limits individual seed growth and nitrogen content. Plant J 62:641–652
Tortell PD (2000) Evolutionary and ecological perspectives on carbon acquisition in phytoplankton. Limnol Oceanogr 45:744–750
Tsuji Y, Suzuki I, Shiraiwa Y (2009) Photosynthetic carbon assimilation in the coccolithophorid Emiliania huxleyi (Haptophyta): evidence for the predominant operation of the C3 cycle and the contribution of beta-carboxylases to the active anaplerotic reaction. Plant Cell Physiol 50:318–329
Tsuji Y, Suzuki I, Shiraiwa Y (2012) Enzymological evidence for the function of a plastid-located pyruvate carboxylase in the Haptophyte alga Emiliania huxleyi: a novel pathway for the production of C4 compounds. Plant Cell Physiol 53:1043–1052
Tsuji Y, Mahardika A, Matsuda Y (2017a) Evolutionarily distinct strategies for the acquisition of inorganic carbon from seawater in marine diatoms. J Exp Bot 68:3949–3958
Tsuji Y, Nakajima K, Matsuda Y (2017b) Molecular aspects of the biophysical CO2-concentrating mechanism and its regulation in marine diatoms. J Exp Bot 68:3763–3772
Voznesenskaya EV, Franceschi VR, Kiirats O, Artyusheva EG, Freitag H, Edwards GE (2002) Proof of C4 photosynthesis without Kranz anatomy in Bienertia cycloptera (Chenopodiaceae). Plant J 31:649–662
Wang Y, Bräutigam A, Weber APM, Zhu XG (2014) Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J Exp Bot 65:3567–3578
Weber T, Gruber A, Kroth PG (2009) The presence and localization of thioredoxins in diatoms, unicellular algae of secondary endosymbiotic origin. Mol Plant 2:468–477
Wheeler MCG, Arias CL, Maurino VG, Andreo CS, Drincovich MF (2009) Identification of domains involved in the allosteric regulation of cytosolic Arabidopsis thaliana NADP-malic enzymes. Febs J 276:5665–5677
Wynn JP, Ratledge C (1997) Malic enzyme is a major source of NADPH for lipid accumulation by Aspergillus nidulans. Microbiology 143:253–257
Wynn JP, Kendrick A, Ratledge C (1997) Sesamol as an inhibitor of growth and lipid metabolism in Mucor circinelloides via its action on malic enzyme. Lipids 32:605–610
Yang ZK, Niu YF, Ma YH, Xue J, Zhang MH, Yang WD, Liu JS, Lu SH, Guan YF, Li HY (2013) Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol Biofuels 6:67:1–14
Young JN, Hopkinson BM (2017) The potential for co-evolution of CO2-concentrating mechanisms and Rubisco in diatoms. J Exp Bot 68:3751–3762
Young JN, Heureux AM, Sharwood RE, Rickaby RE, Morel FM, Whitney SM (2016) Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms. J Exp Bot 67:3445–3456
Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE (2000) Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J Phycol 36:379–386
Zhang Y, Adams IP, Ratledge C (2007) Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153:2013–2025
Zhang YZ, Yin LY, Jiang HS, Li W, Gontero B, Maberly SC (2014) Biochemical and biophysical CO2 concentrating mechanisms in two species of freshwater macrophyte within the genus Ottelia (Hydrocharitaceae). Photosynth Res 121:285–297
Acknowledgements
We are grateful to D. Ballert (Universität Konstanz) for the genetic transformation of P. tricornutum and the cultivation of the transformed cell lines; to the Bioimaging Center at the University of Konstanz for providing the LSM 510 META; to Dr. Katsura Izui for providing the DCDP and to Dr. Pavel Hrouzek (Centre Algatech) for providing the MitoTracker. This study was supported by the German-Israel Foundation for Scientific Research and Development (GIF; Project No. G-1011-199.12/2008 to A.K. and P.G.K.); the University of Konstanz, the Grant-in-Aid for Scientific Research B (Grant No. 24310015 to Y. M.); the Grant-in-Aid for Scientific Research on Innovative Areas (Grant No. 16H06557 to Y. M.); the Japan Society for the Promotion of Science (JSPS; Grant No. 26870750 to S. K.) and by a 2 years Postdoctoral Researcher Fellowship (Mzdová podpora postdoktorandů) from the Academy of Sciences of the Czech Republic (to D.E.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11120_2018_500_MOESM1_ESM.tif
Online Resource Figure 5—Additional images of proteins which subcellular localization has been experimentally proven in a second and independent laboratory. Presented are fluorescence microscopy images showing transformed P. tricornutum cells with expressed protein::GFP fusion proteins of PEPCK, PPDK and ME1. Bright field, chlorophyll auto-fluorescence (red), GFP fluorescence (green) and a merged image, showing an overlap of the chlorophyll auto- and GFP fluorescence, are shown from left to right. Scale bars of each image represent 5 µm. “full” or “pre” suffixes after the enzyme’s name indicate whether a full length or pre-sequence was used for plasmid construction. PEPCK and ME1 are located in the mitochondrion and PPDK is located in the cytosol. P. tricornutum strain UTEX 642 was used to generate the PPDKfull and strain UTEX 646 was used to generate the PEPCKpre and ME1pre transformed cell lines. Abbreviations: PEPCK: phosphoenolpyruvate carboxykinase; PPDK: pyruvate phosphate dikinase; ME: malic enzyme. (TIF 37800 KB)
11120_2018_500_MOESM3_ESM.xlsx
Online Resource Table V—A list of enzymes including protein IDs and localizations that are mentioned in this study. “Target P” (Emanuelsson et al. 2000) shows the highest probability for the predicted subcellular localization. The chosen organism groups are “Non-plant” for P. tricornutum and “Plant” for A. thaliana. The output style is: cTP - chloroplast transit peptide; mTP - mitochondrial transit peptide; SP - secretory pathway, other - cytosol, “cleavage site” indicates the sequence motif surrounding the signal peptide cleavage site predicted by SignalP 3.0 NN (Bendtsen et al. 2004) (*indicates manual predictions of the putative cleavage site motif), “localization experiment” indicates the results of experimental localizations cited in “reference/comments”. (XLSX 37 KB)
Rights and permissions
About this article
Cite this article
Ewe, D., Tachibana, M., Kikutani, S. et al. The intracellular distribution of inorganic carbon fixing enzymes does not support the presence of a C4 pathway in the diatom Phaeodactylum tricornutum. Photosynth Res 137, 263–280 (2018). https://doi.org/10.1007/s11120-018-0500-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-018-0500-5