Abstract
NAC (NAM, ATAF1/2, and CUC2) transcription factors play important roles in the process of abiotic stress response in plants. However, little is known about the functional roles of NACs in Picea (P.) wilsonii. In this study, we functionally characterized a novel P. wilsonii NAC transcription factor PwNAC30 by heterologous expression in Arabidopsis. The results showed that PwNAC30 is mainly localized in the nucleus by transient expression in Nicotiana benthamiana and can function as a nuclear localization transcription factor. β-Glucuronidase (GUS) staining in transgenic Arabidopsis (PwNAC30-promoter-GUS) confirmed the expression pattern of PwNAC30 throughout the plant development process, which is expressed in most of the plant tissues except stamens and petals. Overexpression of PwNAC30 in Arabidopsis significantly repressed the tolerance of seedlings and mature plants to both drought and salt stresses, while exerting no influence on growth and development of plants. The accumulation of reactive oxygen species (ROS) was significantly increased in transgenic plants, and the expression of some stress-responsive genes was obviously inhibited in PwNAC30 overexpression lines. Our study reveals that PwNAC30 functions as a negative regulator of plant tolerance to drought or salt stress, which provides new insights into abiotic tolerance mechanisms in woody plants.











Similar content being viewed by others
References
An JP, Li R, Qu FJ, You CX, Wang XF, Hao YJ (2018) An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J Plant Physiol 221:74–80. https://doi.org/10.1016/j.jplph.2017.12.009
An JP, Yao JF, Xu RR, You CX, Wang XF, Hao YJ (2019) An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response. Physiol Plant 166:472–472. https://doi.org/10.1111/ppl.12695
Baxter A, Mittler R, Suzuki N (2014) ROS as key players in plant stress signalling. J Exp Bot 65:1229–1240. https://doi.org/10.1093/jxb/ert375
Deng YW, Zhai K, Xie Z, Yang D, Zhu X, Liu J, Wang X, Qin P, Yang Y, Zhang G, Li Q, Zhang J, Wu S, Milazzo J, Mao B, Wang E, Xie H, Tharreau D, He Z (2017) Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355:962–965. https://doi.org/10.1126/science.aai8898
Ding AQ et al (2019) RhNAC31, a novel rose NAC transcription factor, enhances tolerance to multiple abiotic stresses in Arabidopsis. Acta Physiologiae Plantarum 41(6):75. https://doi.org/10.1007/s11738-019-2866-1
Fang YJ, You J, Xie KB, Xie WB, Xiong LZ (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Gen Genomics 280:547–563. https://doi.org/10.1007/s00438-008-0386-6
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LSP, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876. https://doi.org/10.1111/j.1365-313X.2004.02171.x
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930. https://doi.org/10.1016/j.plaphy.2010.08.016
Guilfoyle TJ (1997) The structure of plant gene promoters. Genet Eng 19. https://doi.org/10.1007/978-1-4615-5925-2_2
Guo P, Li Z, Huang P, Li B, Fang S, Chu J, Guo H (2017) A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29:2854–2870. https://doi.org/10.1105/tpc.17.00438
Han DG, Hou YJ, Wang YF, Ni BX, Li ZT, Yang GH (2019) Overexpression of a Malus baccata WRKY transcription factor gene (MbWRKY5) increases drought and salt tolerance in transgenic tobacco. Can J Plant Sci 99:173–183. https://doi.org/10.1139/cjps-2018-0053
Hao YJ, Song QX, Chen HW, Zou HF, Wei W, Kang XS, Ma B, Zhang WK, Zhang JS, Chen SY (2010) Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232:1033–1043. https://doi.org/10.1007/s00425-010-1238-2
Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, Zhang JS, Chen SY (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant Journal for Cell & Molecular Biology 68:302–313. https://doi.org/10.1111/j.1365-313x.2011.04687.x
He K, Zhao X, Chi X, Wang Y, Jia C, Zhang H, Zhou G, Hu R (2019) A novel Miscanthus NAC transcription factor MlNAC10 enhances drought and salinity tolerance in transgenic Arabidopsis. J Plant Physiol 233:84–93. https://doi.org/10.1016/j.jplph.2019.01.001
Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13–13. https://doi.org/10.1186/1746-4811-1-13
Hu P, Zhang KM, Yang CP (2019) BpNAC012 positively regulates abiotic stress responses and secondary wall biosynthesis. Plant Physiol 179:700–717. https://doi.org/10.1104/pp.18.01167
Huang L, Hong YB, Zhang HJ, Li DY, Song FM (2016) Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. Bmc Plant Biology 16:203. https://doi.org/10.1186/s12870-016-0897-y
Jensen MK, Hagedorn PH, de Torres-Zabala M, Grant MR, Rung JH, Collinge DB, Lyngkjaer MF (2008) Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp hordei in Arabidopsis. Plant J 56:867–880. https://doi.org/10.1111/j.1365-313X.2008.03646.x
Jin C et al (2017) A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Frontiers Plant Sci 8:1049. https://doi.org/10.3389/fpls.2017.01049
Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, Zhu JK (2006) The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci U S A 103:18816–18821. https://doi.org/10.1073/pnas.0604711103
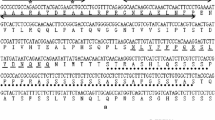
Kehao L, Yongjiang S, Yihang Y, Lingyun Z (2019) Cloning and analysis of a transcription factor PwNAC30 and the promoter sequence in Picea wilsonii. Acta Botanica Boreali-Occidentalia Sinica 39:17–28 CNKI:SUN:DNYX.0.2019-01-002
Kurepin LV, Dahal K, Savitch L, Singh J, Bode R, Ivanov A, Hurry V, Hüner N (2013) Role of CBFs as integrators of chloroplast redox, phytochrome and plant hormone signaling during cold acclimation. Int J Mol Sci 14:12729–12763. https://doi.org/10.3390/ijms140612729
Lee DK, Chung PJ, Jeong JS, Jang G, Bang SW, Jung H, Kim YS, Ha SH, Choi YD, Kim JK (2017) The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol J 15:754–764. https://doi.org/10.1111/pbi.12673
Li S, Wang N, Ji D, Zhang W, Wang Y, Yu Y, Zhao S, Lyu M, You J, Zhang Y, Wang L, Wang X, Liu Z, Tong J, Xiao L, Bai MY, Xiang F (2019) A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell 31:9–2130. https://doi.org/10.1105/tpc.18.00662
Liu B et al (2014a) Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. Plos One 9:7. ARTN e102067. https://doi.org/10.1371/journal.pone.0102067
Liu J, Xu X, Xu Q, Wang SH, Xu JC (2014b) Transgenic tobacco plants expressing PicW gene from Picea wilsonii exhibit enhanced freezing tolerance. Plant Cell Tiss Org 118:391–400. https://doi.org/10.1007/s11240-014-0491-7
Liu C, Wang BM, Li ZX, Peng ZH, Zhang JR (2018a) TsNAC1 is a key transcription factor in abiotic stress resistance and growth. Plant Physiol 176:742–756. https://doi.org/10.1104/pp.17.01089
Liu Q, Yan S, Huang W, Yang J, Dong J, Zhang S, Zhao J, Yang T, Mao X, Zhu X, Liu B (2018b) NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol Biol 98:289–302. https://doi.org/10.1007/s11103-018-0768-z
Liu Y, Wei H, Ma M, Li Q, Kong D, Sun J, Ma X, Wang B, Chen C, Xie Y, Wang H (2019) Arabidopsis FHY3 and FAR1 proteins regulate the balance between growth and defense responses under shade conditions. Plant Cell 31:2089–2106. https://doi.org/10.1105/tpc.18.00991
Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63:289–305. https://doi.org/10.1007/s11103-006-9089-8
Meng XX, Li L, de Clercq I, Narsai R, Xu Y, Hartmann A, Claros DL, Custovic E, Lewsey MG, Whelan J, Berkowitz O (2019) ANAC017 coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol 180:634–653. https://doi.org/10.1104/pp.18.01603
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell and Environment 33:453–467. https://doi.org/10.1111/j.1365-3040.2009.02041.x
Moore K, Roberts LJ (1998) Measurement of lipid peroxidation. Free Radical Research Communications 28:659–671. https://doi.org/10.3109/10715769809065821
Nakashima A, Chen L, Thao NP, Fujiwara M, Wong HL, Kuwano M, Umemura K, Shirasu K, Kawasaki T, Shimamoto K (2008) RACK1 functions in rice innate immunity by interacting with the rac1 immune complex. Plant Cell 20:2265–2279. https://doi.org/10.1105/tpc.107.054395
Nguyen KH, Mostofa MG, Li W, van Ha C, Watanabe Y, le DT, Thao NP, Tran LSP (2018) The soybean transcription factor GmNAC085 enhances drought tolerance in Arabidopsis. Environ Exp Bot 151:12–20. https://doi.org/10.1016/j.envexpbot.2018.03.017
Nguyen KH, Mostofa MG, Watanabe Y, Tran CD, Rahman MM, Tran LSP (2019) Overexpression of GmNAC085 enhances drought tolerance in Arabidopsis by regulating glutathione biosynthesis, redox balance and glutathione-dependent detoxification of reactive oxygen species and methylglyoxal. Environ Exp Bot 161:242–254. https://doi.org/10.1016/j.envexpbot.2018.12.021
Oda-Yamamizo C, Mitsuda N, Sakamoto S, Ogawa D, Ohme-Takagi M, Ohmiya A (2016) The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Scientific Reports 6:23609. https://doi.org/10.1038/srep23609
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309. https://doi.org/10.1105/tpc.105.035881
Seo JJ et al (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197. https://doi.org/10.1104/pp.110.154773
Seok H-Y, Woo DH, Nguyen LV, Tran HT, Tarte VN, Mehdi SMM, Lee SY, Moon YH (2016) Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 245:329–341. https://doi.org/10.1007/s00425-016-2609-0
Shapiguzov A et al (2019) Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. Elife 8:e43284. https://doi.org/10.7554/eLife.43284
Shen H, Yin YB, Chen F, Xu Y, Dixon RA (2009) A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenergy Research 2:217–232. https://doi.org/10.1007/s12155-009-9047-9
Shen J, Lv B, Luo L, He J, Mao C, Xi D, Ming F (2017) The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci Rep 7:40641. https://doi.org/10.1038/srep40641
Sukiran N, Ma J, Ma H, Su Z (2019) ANAC019 is required for recovery of reproductive development under drought stress in Arabidopsis. Plant Mol Biol 99:161–174. https://doi.org/10.1007/s11103-018-0810-1
Tiancong Q et al (2015) Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27:1634–1649. https://doi.org/10.1105/tpc.15.00110
Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498. https://doi.org/10.1105/tpc.104.022699
Tsai SJ, Yin MC (2008) Antioxidative and anti-inflammatory protection of oleanolic acid and ursolic acid in PC12 cells. J Food Sci 73:H174–H178. https://doi.org/10.1111/j.1750-3841.2008.00864.x
Wang LQ, Li Z, Lu MZ, Wang YC (2017) ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis. Frontiers in Plant Science 8:635. https://doi.org/10.3389/fpls.2017.00635
Wang JF et al (2018) Overexpression of BoNAC019, a NAC transcription factor from Brassica oleracea, negatively regulates the dehydration response and anthocyanin biosynthesis in Arabidopsis. Sci Rep-Uk 8:13349. https://doi.org/10.1038/s41598-018-31690-1
Wei QH et al (2017) A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt stress responses in transgenic tobacco plants. Frontiers in Plant Science 8:1374. https://doi.org/10.3389/fpls.2017.01374
Wu R, Duan LN, Pruneda-Paz JL, Oh DH, Pound M, Kay S, Dinneny JR (2018) The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation. Plant Physiol 177:1650–1665. https://doi.org/10.1104/pp.18.00401
Wu JD, Jiang YL, Liang YN, Chen L, Chen WJ, Cheng BJ (2019) Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol Biochem 137:179–188. https://doi.org/10.1016/j.plaphy.2019.02.010
Xiong HY, Yu J, Miao J, Li J, Zhang H, Wang X, Liu P, Zhao Y, Jiang C, Yin Z, Li Y, Guo Y, Fu B, Wang W, Li Z, Ali J, Li Z (2018) Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol 178:451–467. https://doi.org/10.1104/pp.17.01492
Xu GY et al (2017) uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545:491. https://doi.org/10.1038/nature22372
Xu XY, Yao XZ, Lu LT, Zhao DG (2018) Overexpression of the transcription factor NtNAC2 confers drought tolerance in tobacco. Plant Mol Biol Report 36:543–552. https://doi.org/10.1007/s11105-018-1096-9
Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23:2155–2168. https://doi.org/10.1105/tpc.111.084913
Yang X, Wang X, Ji L, Yi Z, Fu C, Ran J, Hu R, Zhou G (2015) Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep 34:943–958. https://doi.org/10.1007/s00299-015-1756-2
Yang XW, He K, Chi X, Chai G, Wang Y, Jia C, Zhang H, Zhou G, Hu R (2018) Miscanthus NAC transcription factor MlNAC12 positively mediates abiotic stress tolerance in transgenic Arabidopsis. Plant Sci 277:229–241. https://doi.org/10.1016/j.plantsci.2018.09.013
Yong Y, Zhang Y, Lyu Y (2019) A stress-responsive NAC transcription factor from tiger lily (LlNAC2) interacts with LlDREB1 and LlZHFD4 and enhances various abiotic stress tolerance in Arabidopsis. Int J Mol Sci 20:3225. https://doi.org/10.3390/ijms20133225
Yu YL, Li Y, Huang G, Meng Z, Zhang D, Wei J, Yan K, Zheng C, Zhang L (2011) PwHAP5, a CCAAT-binding transcription factor, interacts with PwFKBP12 and plays a role in pollen tube growth orientation in Picea wilsonii. J Exp Bot 62:4805–4817. https://doi.org/10.1093/jxb/err120
Yuan X, Wang H, Cai JT, Bi Y, Li DY, Song FM (2019) Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. Bmc Plant Biology 19(1):278. https://doi.org/10.1186/s12870-019-1883-y
Zhang T, Zhang D, Liu Y, Luo C, Zhou Y, Zhang L (2015) Overexpression of a NF-YB3 transcription factor from Picea wilsonii confers tolerance to salinity and drought stress in transformed Arabidopsis thaliana. Plant Physiology & Biochemistry 94:153–164. https://doi.org/10.1016/j.plaphy.2015.05.001
Zhang S, Li C, Wang R, Chen Y, Shu S, Huang R, Zhang D, Li J, Xiao S, Yao N, Yang C (2017) The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol 173:2294–2307. https://doi.org/10.1104/pp.16.00008
Zhang B, Guo GX, Lu F, Song Y, Liu Y, Xu JC, Gao W (2018a) PicW2 from Picea wilsonii: preparation, purification, crystallization and X-ray diffraction analysis. Acta Crystallogr F 74:363–366. https://doi.org/10.1107/S2053230x18007537
Zhang HH, Cui XY, Guo YX, Luo CB, Zhang LY (2018b) Picea wilsonii transcription factor NAC2 enhanced plant tolerance to abiotic stress and participated in RFCP1-regulated flowering time. Plant Mol Biol 98:471–493. https://doi.org/10.1007/s11103-018-0792-z
Zhang XW, Chen LT, Wang JR, Wang MH, Yang SL, Zhao CM (2018c) Photosynthetic acclimation to long-term high temperature and soil drought stress in two spruce species (Picea crassifolia and P-wilsonii) used for afforestation. J For Res 29:363–372. https://doi.org/10.1007/s11676-017-0468-6
Zhang XM, Cheng ZH, Zhao K, Yao WJ, Sun XM, Jiang TB, Zhou BR (2019) Functional characterization of poplar NAC13 gene in salt tolerance. Plant Sci 281:1–8. https://doi.org/10.1016/j.plantsci.2019.01.003
Zhu B, Huo DA, Hong XX, Guo J, Peng T, Liu J, Huang XL, Yan HQ, Weng QB, Zhang XC, du XY (2019a) The Salvia miltiorrhiza NAC transcription factor SmNAC1 enhances zinc content in transgenic Arabidopsis. Gene 688:54–61. https://doi.org/10.1016/j.gene.2018.11.076
Zhu Z, Li G, Yan C, Liu L, Zhang Q, Han Z, Li B (2019b) DRL1, encoding a NAC transcription factor, is involved in leaf senescence in grapevine. Int J Mol Sci 20:11. https://doi.org/10.3390/ijms20112678
Acknowledgments
We thank Professor Dapeng Zhang (College of Life Sciences, Tsinghua University) for providing the pCAMBIA1205 vector and pBI121 vector.
Funding
This research was funded by a grant from the Agricultural Ministry of China, grant number 2016ZX08009-003.
Author information
Authors and Affiliations
Contributions
LZ and KL conceived the project and designed the experiments; LZ collected the samples; KL, YY, YM, and AW performed the experiments; KL conducted the data analysis and drafted the manuscript; LZ revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
The function of PwNAC30, which is identified from Picea wilsonii, is characterized by heterologous expression in Arabidopsis. Our results demonstrated that PwNAC30, as a nuclear localization transcription factor, plays a negative role in drought and salt stress response in plants; moreover, the inhibition mechanism of tolerance to abiotic stress in transgenic plants is achieved by the accumulation of reactive oxygen species (ROS) and the downregulation of stress-related genes.
Electronic Supplementary Material
Table S1
(DOCX 17 kb)
Fig. S1
Detection of PwNAC30 in transcription Arabidopsis. (a) Validation of PwNAC30 by RT-PCR. (b) The relative expression level of PwNAC30 in transgenic plants. WT, wild-type; NC, plant lines transformed with empty vectors; OE-2 and OE-4, transgenic lines of PwNAC30 overexpression. For qRT-PCR, AtActin was used as reference gene. Each value is the mean value ± SE of three independent determinations, and different letters indicate significant differences at P < 0.05 by Duncan’s multiple range test (PNG 1640 kb)
Fig. S2
Growth characteristics of PwNAC30 overexpression lines. (a, e) Plant heights of OE lines, WT and NC plants under normal growth condition. (b) Detached rosette leaves of OE lines, WT and NC plants under normal growth condition. (c) Flower organs of OE lines, WT and NC plants under normal growth condition. (d, f) Maturing siliques of OE lines, WT and NC plants. (g) Flowering times of OE lines, WT and NC plants under normal growth condition (WT, wild-type; NC, plant lines transformed with empty vectors; OE-2 and OE-4, transgenic lines of PwNAC30 overexpression). Each value is the mean value ± standard error of three independent determinations, and different letters indicate significant differences at P < 0.05 by Duncan’s multiple range test (PNG 6033 kb)
Fig. S3
Effects of exogenous ABA on the activity of the promoter. (a) GUS activity driven by the promoter of PwNAC30 under different concentrations of exogenous ABA. (b) Analysis of the PwNAC30 promoter under different ABA treatments through dual-luciferase reporter assay (PC, 35S-GUS, empty pBI121 vector transformed into N. benthamiana leaves; NC, N. benthamiana injected with buffer solution). Each value is the mean value ± standard error of three independent determinations, and different letters indicate significant differences at P < 0.05 by Duncan’s multiple range test (PNG 4144 kb)
Rights and permissions
About this article
Cite this article
Liang, Kh., Wang, Ab., Yuan, Yh. et al. Picea wilsonii NAC Transcription Factor PwNAC30 Negatively Regulates Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Mol Biol Rep 38, 554–571 (2020). https://doi.org/10.1007/s11105-020-01216-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01216-z