Abstract
Cabbage (Brassica oleracea var. capitata) is the most popular leafy vegetable; however, its quality as a vegetable depends on its growth stage. Premature bolting triggered by low temperatures leads to a reduction of yield and quality of cabbage. Late bolting is preferred by growers to increase market value, whereas early bolting plants are ideal for quality seed production. Herein, we reported a gene BolPrx.2 annotated as Q9FLC0 in the SwissPort, involved in bolting time variation in cabbage and designed molecular markers to characterize early- and late-bolting cabbage populations and lines. The BolPrx.2 gene encodes a peroxidase domain and has been identified as a candidate showed almost similar effect as the previously reported MADS-box domain-containing FLC genes for controlling bolting time. An insertion/deletion (InDel) variation in intron1 has been identified as a causal factor for variation between late- and early-bolting inbred lines. By using this InDel, we designed molecular markers for characterizing the bolting time variation and validated them with 141 F2 generation plants. These markers predicted about 84% of the variation within the population and commercial lines. Therefore, it could be a potential genetic tool to predict bolting time variation and support marker-assisted back crossing (MABC) programs for developing desired bolting types of cabbage cultivars.








Similar content being viewed by others
References
Anderson JT, Lee CR, Mitchell-Olds T (2011) Life-history QTLS and natural selection on flowering time in Boechera stricta, a perennial relative of Arabidopsis. Evolution 65:771–787
Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13:627–639
Badiani M, Paolacci A, Miglietta F, Kimball B, Pinter P, Garcia R, Hunsaker D, LaMorte R, Wall G (1996) Seasonal variations of antioxidants in wheat (Triticum aestivum) leaves grown under field conditions. Funct Plant Biol 23:687–698
Blázquez MA, Green R, Nilsson O, Sussman MR, Weigel D (1998) Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10:791–800
Camargo L, Osborn T (1996) Mapping loci controlling flowering time in Brassica oleracea. Theor Appl Genet 92:610–616
Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43:956–963
Chai L, Wang J, Fan Z, Liu Z, Wen G, Li X, Yang Y (2012) Regulation of the flowering time of Arabidopsis thaliana by thylakoid ascorbate peroxidase. Afr J Biotechnol 11:7151–7157
Consortium EUAGS (2000) Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408:823
Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141:550–550. e552
Franks SJ, Perez-Sweeney B, Strahl M, Nowogrodzki A, Weber JJ, Lalchan R, Jordan KP, Litt A (2015) Variation in the flowering time orthologs BrFLC and BrSOC1 in a natural population of Brassica rapa. PeerJ 3:e1339
Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132:1107–1114
Gielis J, Goetghebeur P, Debergh P (1999) Physiological aspects and experimental reversion of flowering in Fargesia murieliae (Poaceae, Bambusoideae). Syst Geogr Plants 68:147–158
Gómez-Mena C, Piñeiro M, Franco-Zorrilla JM, Salinas J, Coupland G, Martínez-Zapater JM (2001) Early bolting in short days: an Arabidopsis mutation that causes early flowering and partially suppresses the floral phenotype of leafy. Plant Cell 13:1011–1024
Gotô N, Hamada M (1988) Promotion of flowering by DNA base analogues and changes in acid phosphatase and peroxidase isozyme composition in dark-grown Arabidopsis thaliana. Plant Cell Physiol 29:683–688
Grillo MA, Li C, Hammond M, Wang L, Schemske DW (2013) Genetic architecture of flowering time differentiation between locally adapted populations of Arabidopsis thaliana. New Phytol 197:1321–1331
Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, Von Korff M, Varshney RK, Graner A, Valkoun J (2009) Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot 60:3531–3544
Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21:4327–4337
Hirai N, Kojima Y, Shinozaki M, Koshimizu K, Murofushi N, Takimoto A (1995) Accumulation of ascorbic acid in the cotyledons of morning glory (Pharbitis nil) seedlings during the induction of flowering by low-temperature treatment and the effect of prior exposure to high-intensity light. Plant Cell Physiol 36:1265–1271
Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J-F, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43:476–481
Ishizawa M, Kobayashi Y, Miyamura T, Matsuura S (1991) Simple procedure of DNA isolation from human serum. Nucleic Acids Res 19:5792
Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290:344–347
Jung C, Müller AE (2009) Flowering time control and applications in plant breeding. Trends Plant Sci 14:563–573
Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55:521–535
Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart C, Peeters A (1998a) Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148:885–892
Koornneef M, Alonso-Blanco C, Peeters AJ, Soppe W (1998b) Genetic control of flowering time in Arabidopsis. Annu Rev Plant Biol 49:345–370
Krekule J, Machackova I (1986) possible role of auxin and of its metabolic changes in the photoperiodic control of flowering. Molecular and physiological aspects of plant peroxidases/edited by H Greppin, C Penel, Th Gaspar. Information Systems Division, National Agricultural Library, Beltsville, Maryland and Washington, D.C., USA
Levy YY, Dean C (1998) The transition to flowering. Plant Cell 10:1973–1989
Li Y, Roycewicz P, Smith E, Borevitz JO (2006) Genetics of local adaptation in the laboratory: flowering time quantitative trait loci under geographic and seasonal conditions in Arabidopsis. PLoS One 1:e105
Li P, Filiault D, Box MS, Kerdaffrec E, van Oosterhout C, Wilczek AM, Schmitt J, McMullan M, Bergelson J, Nordborg M (2014) Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev 28:1635–1640
Lokhande SD, Ogawa KI, Tanaka A, Hara T (2003) Effect of temperature on ascorbate peroxidase activity and flowering of Arabidopsis thaliana ecotypes under different light conditions. J Plant Physiol 160:57–64
Mao Y, Wu F, Yu X, Bai J, Zhong W, He Y (2014) MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in Chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol 164:710–720
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Michaels SD, Amasino RM (2000) Memories of winter: vernalization and the competence to flower. Plant Cell Environ 23:1145–1153
Michaels SD, He Y, Scortecci KC, Amasino RM (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci U S A 100:10102–10107
Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137:149–156
Moharekar S, Tanaka R, Ogawa K, Tanaka A, Hara T (2007) Great promoting effect of high irradiance from germination on flowering in Arabidopsis thaliana—a process of photo-acclimation. Photosynthetica 45:259–265
Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39:877–885
Ogawa KI, Tasaka Y, Mino M, Tanaka Y, Iwabuchi M (2001) Association of glutathione with flowering in Arabidopsis thaliana. Plant Cell Physiol 42:524–530
Österberg MK, Shavorskaya O, Lascoux M, Lagercrantz U (2002) Naturally occurring indel variation in the Brassica nigra COL1 gene is associated with variation in flowering time. Genetics 161:299–306
Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL (2001) Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol 126:122–132
Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15:1159–1169
Razi H, Howell E, Newbury H, Kearsey M (2008) Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor Appl Genet 116:179–192
Ridge S, Brown PH, Hecht V, Driessen RG, Weller JL (2014) The role of BoFLC2 in cauliflower (Brassica oleracea var. botrytis L.) reproductive development. J Exp Bot 66:125–135
Ross J, O'Neill D (2001) New interactions between classical plant hormones. Trends Plant Sci 6:2–4
Saha G, Park J-I, Jung H-J, Ahmed NU, Kayum MA, Chung M-Y, Hur Y, Cho Y-G, Watanabe M, Nou I-S (2015) Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genomics 16:178
Salomé PA, Bomblies K, Laitinen RA, Yant L, Mott R, Weigel D (2011) Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188:421–433
Sánchez-Bermejo E, Méndez-Vigo B, Pico FX, Martínez-Zapater JM, Alonso-Blanco C (2012) Novel natural alleles at FLC and LVR loci account for enhanced vernalization responses in Arabidopsis thaliana. Plant Cell Environ 35:1672–1684
Schiessl SV, Huettel B, Kuehn D, Reinhardt R, Snowdon RJ (2017) Flowering time gene variation in Brassica species shows evolutionary principles. Front Plant Sci 8:1742
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Schranz ME, Quijada P, Sung S-B, Lukens L, Amasino R, Osborn TC (2002) Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162:1457–1468
Scortecci KC, Michaels SD, Amasino RM (2001) Identification of a MADS-box gene, FLOWERING LOCUS M that represses flowering. Plant J 26:229–236
Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20:898–912
Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11:445–458
Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci U S A 97:3753–3758
Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14:2527–2537
Shigeto J, Tsutsumi Y (2016) Diverse functions and reactions of class III peroxidases. New Phytol 209:1395–1402
Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138:1163–1173
Song X, Duan W, Huang Z, Liu G, Wu P, Liu T, Li Y, Hou X (2015) Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom. Sci Rep 5:14631
Strange A, Li P, Lister C, Anderson J, Warthmann N, Shindo C, Irwin J, Nordborg M, Dean C (2011) Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS One 6:e19949
Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM (2006) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38:706–710
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J-H, Bancroft I, Cheng F (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Wang Y, Wu F, Bai J, He Y (2014) BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol J 12:312–321
Weigel D, Meyerowitz EM (1993) Activation of floral homeotic genes in Arabidopsis. Science 261:1723–1726
Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D (2005) FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170:1197–1207
Xia G, He Q, Zhao S (2015) Physiological and biochemical properties analysis of late-bolting transgenic Chinese cabbage (Brassica rapa L. ssp. pekinensis). J Anim Plant Sci 25:152–157
Zang YX, Kim HU, Kim JA, Lim MH, Jin M, Lee SC, Kwon SJ, Lee SI, Hong JK, Park TH (2009) Genome-wide identification of glucosinolate synthesis genes in Brassica rapa. FEBS J 276:3559–3574
Zou X, Suppanz I, Raman H, Hou J, Wang J, Long Y, Jung C, Meng J (2012) Comparative analysis of FLC homologues in Brassicaceae provides insight into their role in the evolution of oilseed rape. PLoS One 7:e45751
Funding
This research was supported by the Golden Seed Project (Center for Horticultural Seed Development, No. 213007-05-2-SB110), the Ministry of Agriculture, the Food and Rural Affairs (MAFRA), the Ministry of Oceans and Fisheries (MOF), the Rural Development Administration (RDA), and Korea Forest Service (KFS), Korea.
Author information
Authors and Affiliations
Contributions
The work presented here was carried out in collaboration among all authors. MA carried out the experiments and prepared the tables, figures, and manuscript draft. UKN collected primary data regarding genes and wrote, edited, and finalized the manuscript. JIP formulated the experimental concept and provided the plant materials. HTK conducted greenhouse experiments and took care of the plant populations. MKB performed in silico analyses and helped with cloning. ISN designed and participated in all the experiments and assisted in improving the technical sites for the project. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
ESM 1
Supplementary Table S1. Primer list of 8 genes of BolFLC, BolPrxs, BolATL73 and BolCYP. Supplementary Table S2. Nucleotide and amino acid similarities of BolPrx.2 gene with reported FLC gene and other flowering pathway genes in Brassica oleracea, respectively. (DOC 89 kb)
Supplementary Fig. S1
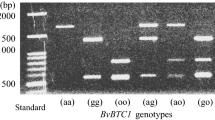
Validation of 20 commercial inbred early- and late-bolting cabbage lines with InDel marker of BolPrx.2 gene matched 85%, the heterozygous lines (lanes: 10, 13, and 16) are not matched with the phenotypes. Here, P1= late-bolting parent (BN4730), P2= early-bolting parent (BN7115), F1= (BN4730 × BN7115), M=100 bp DNA marker, red color numbered (10 early-bolting lines): lane 1, 2, 4, 7, 9, 11, 12, 15,17 are 17FLE1, 17FLE2, 17FLE3, 17FLE4, 17FLE5, 17FLE6, 17FLE7, 17FLE8, 17FLE9, and 17FLE10, respectively and black color numbered (10 late-bolting lines): lanes 3, 5, 6, 8, 10, 13, 14, 16 ,19 and 20 are 17FLL1, 17FLL2, 17FLL3, 17FLL4, 17FLL5, 17FLL6, 17FLL7, 17FLL8, 17FLL9, and 17FLL10, respectively. (PPT 493 kb)
Additional file 1.
List of the genes and their sequences used for constructing of phylogenetic tree. (DOC 46 kb)
Additional file 2.
Sequence alignment of the total sequences of the gene BolPrx.2 cloned from late bolting line (BN4730) and early bolting line (BN7115) aligned with reference sequence of BolPrx.2 gene collected from Brassica database (http://brassicadb.org/brad/index.php). Blue and black colour regions are the introns and exons of the gene, respectively. Gray highlighted codons indicate the start and stop codon of the gene; yellow, pink, and green highlighted portion indicate the deletion, insertion and SNPs variations in the late bolting line. (DOC 41 kb)
Rights and permissions
About this article
Cite this article
Abuyusuf, M., Nath, U.K., Kim, HT. et al. Intronic Sequence Variations in a Gene with Peroxidase Domain Alter Bolting Time in Cabbage (Brassica oleracea var. capitata). Plant Mol Biol Rep 36, 725–737 (2018). https://doi.org/10.1007/s11105-018-1113-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-018-1113-z