Abstract
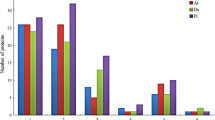
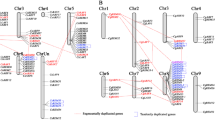
MADS-box genes encode transcription factors that are involved in developmental control and signal transduction in plants, animals and fungi. To date, few citrus MADS-box genes and limited tissue expression profiling have been reported. In this study, 84 clementine mandarin and 52 sweet orange MADS-box genes have been identified using bioinformatics analysis. Subsequently, comparative analysis of these genes, including gene structures, phylogenetic relationships, conserved protein motifs, expression patterns, and simple sequence repeat (SSR) markers were identified in these citrus species. According to these analyses, these 136 MADS-box genes can be categorized into five (MIKCC, MIKC*, Mα, Mβ, and Mγ) groups, and the MIKCC group was further divided into 13 subfamilies in two citrus species. In addition, gene duplication of MADS-box genes was also investigated in the clementine mandarin, sweet orange, rice and Arabidopsis genomes. A lower percentage of MADS-box gene duplication was found in the clementine mandarin and sweet orange genomes as compared with Arabidopsis. A survey of the two citrus species based on organ-specific expression profiling indicated that MADS-box genes exhibit various expression patterns, suggesting diverse and novel functions of MADS-box gene families in citrus. In addition, 128 new putative microsatellites with flanking sequences sufficient for primer design were also identified from 136 MADS genes. Sixteen SSRs were selected randomly for validation and transferability testing in 11 citrus species. Among 16 SSRs, 15 were identified as true-to-type SSR loci and were highly transferable among citrus species. These results provide a useful reference for the selection of candidate MADS-box genes for cloning and further functional analysis in citrus.




Similar content being viewed by others
References
Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, De Pouplana LR, Martnez-Castilla L, Yanofsky MF (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97:5328–5333
Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242
Bailey TL, Elkan C (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3:21–29
Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119:721–743
Brunner AM, Nilsson O (2004) Revisiting tree maturation and floral initiation in the poplar functional genomics era. New Phytol 164:43–51
Caetano-Anollés G (2001) Novel strategies to study the role of mutation and nucleic acid structure in evolution. Plant Cell Tissue Organ Cult 67:115–132
Chalhoub BA, Thibault S, Laucou V, Rameau C, Höfte H, Cousin R (1997) Silver staining and recovery of AFLP amplification products on large denaturing polyacrylamide gels. Biotechniques 22:216–220
Chen C, Zhou P, Choi YA, Huang S, Gmitter FG (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257
Chen C, Bowman KD, Choi YA, Dang PM, Rao MN, Huang S, Soneji JR, McCollum TG, Gmitter FG (2008) EST-SSR genetic maps for Citrus sinensis and Poncirus trifoliata. Tree Genet Genomes 4:1–10
Cheng YJ, Guo WW, Yi HL, Pang XM, Deng X (2003) An efficient protocol for genomic DNA extraction from citrus species. Plant Mol Biol Rep 21:177–178
Cho S, Jang S, Chae S, Chung KM, Moon YH, An G, Jang SK (1999) Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol Biol 40:419–429
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37
De Bodt S, Raes J, Van de Peer Y, Theißen G (2003) And then there were many: MADS goes genomic. Trends Plant Sci 8:475–483
Dornelas MC, Patreze CM, Angenent GC, Immink RGH (2012) MADS: the missing link between identity and growth? Trends Plant Sci 16:89–97
Endo T, Shimada T, Fujii H, Omura M (2006) Cloning and characterization of 5 MADS-box cDNAs isolated from citrus fruit tissue. Sci Hortic 109:315–321
Ferrandiz C, Liljegren SJ, Yanofsky MF (2000) Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289:436
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251
Gmitter FG, Chen C, Machado MA, de Souza AA, Ollitrault P, Froehlicher Y, Shimizu T (2012) Citrus genomics. Tree Genet Genomes 8:611–626
Gojobori T, Li WH, Graur D (1982) Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol 18:360–369
Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8:1548–1560
Gu Q, Ferrandiz C, Yanofsky MF, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125:1509–1517
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29:1023–1026
Henschel K, Kofuji R, Hasebe M, Saedler H, Munster T, Theissen G (2002) Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol Biol Evol 19:801–814
Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21:4327–4337
Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin responsive genes during reproductive development and abiotic stress in rice. FEBS J 276:3148–3162
Kaufmann K, Melzer R, Theissen G (2005) MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347:183–198
Kramer EM, Dorit RL, Irish VF (1998) Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149:765–783
Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF (1970) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. J Comp Physiol Psychol 71:1–25
Liu T, Li Y, Zhang C, Qian Y, Wang Z, Hou X (2012) Overexpression of FLOWERING LOCUSC, isolated from Non-heading Chinese cabbage (Brassica campestris ssp. Chinensis makino), influences fertility in Arabidopsis. Plant Mol Biol Rep 30:1444–1449
Lu S-J, Wei H, Wang Y, Wang H-M, Yang R-F, Zhang X-B, Tu J-M (2012) Overexpression of a transcription factor OsMADS15 modifies plant architecture and flowering time in rice (Oryza sativa L.). Plant Mol Biol Rep 30:1461–1469
Masiero S, Colombo L, Grini PE, Schnittger A, Kater MM (2012) The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 23:865–872
Messenguy F, Dubois E (2003) Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316:1–21
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33:867–874
Morin RD, O’Connor MD, Griffith M (2008) Application of massively parallel sequencing to microRNA profiling. Genome Res 18:610–621
Nam J, Kim J, Lee S, An G, Ma H, Nei M (2004) Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc Natl Acad Sci USA 101:1910–1915
Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, Lepiniec L (2002) The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14:2463–2479
Parenicov L, De Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis new openings to the MADS world. Plant Cell 15:1538–1551
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405:200–203
Pillitteri LJ, Lovatt CJ, Walling LL (2004) Isolation and characterization of LEAFY and APETALA1 homologues from Citrus sinensis L. OsbeckWashington’. J Am Soc Hortic Sci 129:846–856
Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL (2001) Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol 126:122–132
Riechmann JL, Meyerowitz EM (1997) MADS domain proteins in plant development. Biol Chem 378:1079
Riechmann JL, Krizek BA, Meyerowitz EM (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93:4793–4798
Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288:1613–1616
Schmid R, Blaxter ML (2008) annot 8 r: GO, EC and KEGG annotation of EST datasets. BMC Bioinforma 9:180
Shore P, Sharrocks AD (1995) The MADS-box family of transcription factors. Eur J Biochem 229:1–13
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tan FC, Swain SM (2007) Functional characterization of AP3, SOC1 and WUS homologues from citrus (Citrus sinensis). Physiol Plant 131:481–495
Tani E, Polidoros AN, Tsaftaris AS (2007) Characterization and expression analysis of FRUITFULL-and SHATTERPROOF-like genes from peach (Prunus persica) and their role in split-pit formation. Tree Physiol 27:649–659
Tapia-López R, García-Ponce B, Dubrovsky JG, Garay-Arroyo A, Pérez-Ruiz RV, Kim SH, Acevedo F, Pelaz S, Alvarez-Buylla ER (2008) An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol 146:1182–1192
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes: review. Cell 78:203–209
Wilkie JD, Sedgley M, Olesen T (2008) Regulation of floral initiation in horticultural trees. J Exp Bot 59:3215–3228
Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346:35–39
Acknowledgments
This research was supported financially by the National Natural Science Foundation of China (grant nos. 31071777, 31130046, and 31101528), the Fundamental Research Funds for the Central Universities (Program No. 20110146120027 and 2013PY083), and the International Foundation for Science No C/5148-1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hou, XJ., Liu, SR., Khan, M.R.G. et al. Genome-Wide Identification, Classification, Expression Profiling, and SSR Marker Development of the MADS-Box Gene Family in Citrus. Plant Mol Biol Rep 32, 28–41 (2014). https://doi.org/10.1007/s11105-013-0597-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-013-0597-9